4628
A deep transfer learning model for early prediction of cognitive deficits using brain structural connectome data in very preterm infants1The Perinatal Institute and Section of Neonatology, Perinatal and Pulmonary Biology, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, United States, 2Department of Electronic Engineering and Computing Science, University of Cincinnati, Cincinnati, OH, United States, 3Department of Radiology, University of Cincinnati College of Medicine, Cincinnati, OH, United States, 4Imaging Research Center, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, United States, 5Department of Biostatistics, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, United States, 6Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, OH, United States
Synopsis
The high risk of cognitive deficits is a major concern for parents and clinicians caring for premature babies. Early and accurate identification of children at risk is urgently needed for early treatment decision. We propose a deep transfer learning model to predict cognitive deficits at 2 years corrected age using brain structural connectome data obtained at term. The proposed model was able to identify infants at high-risk of later cognitive deficit with an accuracy of 78.5% and an AUC of 0.75. The predicted cognitive scores were significantly correlated with corresponding Bayley-III cognitive scores, with a Pearson’s correlation coefficient of 0.48.
INTRODUCTION
Survival of preterm babies has increased worldwide due to improved perinatal care and technological advances. Neurodevelopmental impairments rather than survival alone has become the main concern in children born premature. 1,2 Early identification of children at risk of later cognitive deficits is urgently needed for early treatment decision during the optimal neuroplasticity window when intervention is likely to be most effective. However, despite extensive effort made in studying neurodevelopment impairments, the exact etiology leading to future cognitive deficits in those patient population remains unknown. Brain structural connectome, derived from diffusion MRI (dMRI), may play a key role in connecting brain development and cognitive performance. 3 Data derived from brain structural connectome are intrinsically complex and very high in dimension, which results in difficulties in designing feature extraction methods and building machine learning prognostic models. Recently, deep learning techniques have shown great promise in prediction tasks particularly using high dimensional data. 4 However, the deep learning models usually require large datasets to train, while available neuroimaging datasets are small and expensive to enrich. 5 Transfer learning is the key to mitigating the insufficient datasets problem in deep learning. 6 In this work, we propose a deep transfer learning neural network model and compare it to a deep learning model without transfer learning to predict cognitive deficits at 2 years corrected age using structural brain connectome data obtained at term-equivalent age in very preterm infants.METHODS
This study included 80 very preterm infants with gestational age at birth mean (standard deviation) of 28 (2.4) weeks. MRI imaging was performed at mean (SD) of 40 (0.6) weeks postmenstrual age. The Nationwide Children's Hospital Institutional Review Board approved this study and written parental informed consent was obtained for every subject. All preterm infants received standardized Bayley Scales of Infant and Toddler Development III test at 2 years corrected age. The Bayley-III cognitive scores are on a scale of 40 to 160, with a mean of 100 and a standard deviation of 15. We classified very preterm infants into at high (31 subjects) vs. low risk (49 subjects) for moderate/severe cognitive deficits using a cutoff value of 90. The obtained dMRI data were preprocessed using FMRIB’s Diffusion Toolbox (in the FMRIB Software Library, FSL, Oxford, UK). Head motion and eddy current artifacts were mitigated by aligning all diffusion images to their B0 image via an affine transformation. The whole brain structure connectome was constructed based on 90 regions of interest (ROIs) defined from a neonatal Automated Anatomical Labeling atlas. 7 The weights of structural connectivities between each pair of ROIs were calculated as the mean fractional anisotropy of each voxel intersecting the tract and then averaged over all tracts between the two ROIs, resulting in a 90 x 90 symmetric adjacency matrix. This was performed using the UCLA Multimodal Connectivity Package. 8Figure 1 shows the schematic diagram of our proposed deep transfer learning model. The model has two phases. First, we implement the 1st to 21st layers from a pre-trained VGG19 model with fixed weights to take the brain connectome data as input and to extract high-level features. 9 These 21 layers include 16 convolutional layers and 5 max pooling layers. For each convolutional layer, VGG19 uses small convolutional filters (3x3) along with a rectified linear unit activation function. Second, we attach additional 2 convolutional layers with [256, 256] neurons, 1 max pooling layer and 2 fully connected layers with [256, 64] neurons to take the outputs from VGG19 to further learn the discriminative features of cognitive deficits. Those two convolutional layers both use the same activation functions and filters as VGG19. Finally, an output layer (a SoftMax function) is used for classification or a linear function is used for regression. We evaluate the prediction performance through 5-fold cross-validation with the metrics of: accuracy, sensitivity, specificity, and area under the receiver operating characteristic curve (AUC) for risk stratification and metrics of Pearson’s correlation coefficient, mean absolute error and standard deviation of absolute error for prediction of cognitive scores.
RESULTS
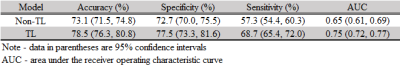
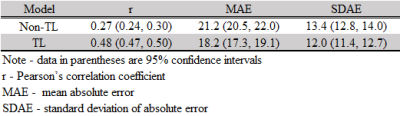
Our model was able to correctly identify subjects with high-risk of cognitive deficits with an accuracy (95% confidence interval) of 78.5% (76.3%, 80.8%) and an AUC of 0.75 (0.72, 0.77) (Table 1). The predicted cognitive scores using our proposed model were significantly correlated with corresponding actual scores, with a Pearson’s correlation coefficient of 0.48 (Table 2).DISCUSSIONS and CONCLUSIONS
Early diagnosis of neurodevelopmental impairments is critical for prevention and treatment efforts. In this work, we developed a deep transfer learning model for the early prediction of cognitive deficits at 2 years of age in very preterm infants using brain structural connectome data. The complexity of structural connectivity data and small sample size make the task challenging. Transfer learning is the key to mitigating the insufficient datasets problem. It takes advantage of knowledge learned from a large dataset and applies it to tasks with small datasets. We demonstrated that the prediction performance of our proposed transfer learning model outperformed the model without transfer learning. Future directions include the incorporation of functional connectivity, anatomical and clinical data with a larger neuroimaging dataset to further improve on the prediction performance.Acknowledgements
This study was supported by the National Institutes of Health grants R21-HD094085, R01-NS094200, and R01-NS096037 and a Trustee grant from Cincinnati Children’s Hospital Medical Center. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.References
1. Jarjour IT. Neurodevelopmental outcome after extreme prematurity: a review of the literature. Pediatr Neurol. 2015;52(2):143-152.
2. Blencowe H, Cousens S, Oestergaard MZ, et al. National, regional, and worldwide estimates of preterm birth rates in the year 2010 with time trends since 1990 for selected countries: a systematic analysis and implications. Lancet. 2012;379(9832):2162-2172.
3. Kawahara J, Brown CJ, Miller SP, et al. BrainNetCNN: Convolutional neural networks for brain networks; towards predicting neurodevelopment. Neuroimage. 2017;146:1038-1049.
4. He L, Li H, Holland SK, Yuan W, Altaye M, Parikh NA. Early prediction of cognitive deficits in very preterm infants using functional connectome data in an artificial neural network framework. NeuroImage: Clinical. 2018;18:290-297.
5. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436-444.
6. Shin HC, Roth HR, Gao M, et al. Deep Convolutional Neural Networks for Computer-Aided Detection: CNN Architectures, Dataset Characteristics and Transfer Learning. IEEE Trans Med Imaging. 2016;35(5):1285-1298.
7. Shi F, Yap PT, Wu G, et al. Infant brain atlases from neonates to 1- and 2-year-olds. PLoS One. 2011;6(4):e18746.
8. Bassett DS, Brown JA, Deshpande V, Carlson JM, Grafton ST. Conserved and variable architecture of human white matter connectivity. Neuroimage. 2011;54(2):1262-1279.
9. Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:14091556. 2014.
Figures