4591
Development of Pediatric Spinal Cord White Matter Atlas: Preliminary Analysis
Shiva Shahrampour1, Benjamin De Leener2, Devon Middleton1, Mahdi Alizadeh1, Laura Krisa1, Adam Flanders3, Scott Faro3, Julien Cohen-Adad2, and Feroze Mohamed1
1Thomas Jefferson University, Philadelphia, PA, United States, 2Polytechnique Montreal, Montreal, QC, Canada, 3Thomas Jefferson University Hospital, Philadelphia, PA, United States
1Thomas Jefferson University, Philadelphia, PA, United States, 2Polytechnique Montreal, Montreal, QC, Canada, 3Thomas Jefferson University Hospital, Philadelphia, PA, United States
Synopsis
Spinal cord diffusion tensor imaging and atlas-based group analysis has potential for improving prognosis of spinal cord related diseases, aid clinical decision making and provide quantitative imaging biomarkers for spinal cord injury assessment.
Purpose:
The purpose of this work is two-folds: 1) To create and develop white matter (WM) atlas of pediatric spinal cord. 2) To obtain atlas based normative values of the diffusion tensor imaging (DTI) parameters for various white matter tracts of the healthy pediatric spinal cord.
Material and Methods:
Diffusion tensor images of 10 healthy subjects (6-16 yrs) were acquired axially at 3T by using 2 overlapping slabs, to cover the cervical and thoracic spinal cord using an inner Field of View (FOV) spin echo based EPI pulse sequence. The imaging parameters of DTI acquisition for each slab is as follow: FOV=164 mm, phase FOV=28.4% (47 mm), 3 averages of 20 diffusion directions, 6 b0 acquisitions, b=800 s/mm2, voxel size= 0.8x0.8x6 mm3, number of slices =40, TR = 7900 ms, TE=110 ms, acquisition time= 8:49 minutes per slab. Generation of the white matter atlas involved the following pipeline: All data were preprocessed using Spinal Cord Toolbox (SCT) [1]. Initially a slice-wise motion correction was performed on all the diffusion images with several degrees of regularization (i.e. smoothing along z axis, outlier detection). The data were then registered to the PAM 50 template incorporated in the SCT toolbox [2]. Next using non-rigid deformations, a series of affine transformations were estimated between the b0 image, the anatomic data and the template. The combined transformations were then used to co-register PAM50 white matter atlas to the patient specific space. The DTI indices of FA (Fractional Anisotropy) and MD (Mean Diffusivity) were then measured for the following specific WM tracts: left and right dorsal fasciculus gracilis, dorsal fasciculus cuneatus and lateral corticospinal (CST). The values were computed at a single slice centered at the C3 vertebral body as shown in figure (1). The mean, standard deviation and variability of the measurements were then compared between tracts using COV (coefficient of variations) in table (1). An ANOVA test was performed to test the effect of laterality (left versus right) and functionality (motor versus sensory).
Results:
Figure (2) shows WM atlas overlaid onto a b0 image. The white matter tracts automatically identified and selected for further quantitative analysis are illustrated in this figure ( green (1): right CST, green (2): left CST, dark blue (3): right fasciculus cuneatus, dark blue (4): left fasciculus cuneatus, red (5): right fasciculus gracilis, red (6): left fasciculus gracilis, yellow: gray matter, light blue: the unlabeled white matter ). Results of ANOVA on the selected tracts using FA show no effect for laterality (p=0.74) and no effect for functionality of tracts (p= 0.85).
Conclusion:
To the best of our knowledge, this work is the first to create WM atlas of pediatric spinal cord. The pipeline incorporates unique post-processing, followed by template registration and quantification of DTI metrics using atlas-based regions. This automatic method eliminates the need for manual region of interest analysis of various WM tracts and therefore increases accuracy of the measurements. Future work with a larger cohort as well as assessing other DTI indices (Radial Diffusivity (RD) and Axial Diffusivity (AD)) is warranted.
The purpose of this work is two-folds: 1) To create and develop white matter (WM) atlas of pediatric spinal cord. 2) To obtain atlas based normative values of the diffusion tensor imaging (DTI) parameters for various white matter tracts of the healthy pediatric spinal cord.
Material and Methods:
Diffusion tensor images of 10 healthy subjects (6-16 yrs) were acquired axially at 3T by using 2 overlapping slabs, to cover the cervical and thoracic spinal cord using an inner Field of View (FOV) spin echo based EPI pulse sequence. The imaging parameters of DTI acquisition for each slab is as follow: FOV=164 mm, phase FOV=28.4% (47 mm), 3 averages of 20 diffusion directions, 6 b0 acquisitions, b=800 s/mm2, voxel size= 0.8x0.8x6 mm3, number of slices =40, TR = 7900 ms, TE=110 ms, acquisition time= 8:49 minutes per slab. Generation of the white matter atlas involved the following pipeline: All data were preprocessed using Spinal Cord Toolbox (SCT) [1]. Initially a slice-wise motion correction was performed on all the diffusion images with several degrees of regularization (i.e. smoothing along z axis, outlier detection). The data were then registered to the PAM 50 template incorporated in the SCT toolbox [2]. Next using non-rigid deformations, a series of affine transformations were estimated between the b0 image, the anatomic data and the template. The combined transformations were then used to co-register PAM50 white matter atlas to the patient specific space. The DTI indices of FA (Fractional Anisotropy) and MD (Mean Diffusivity) were then measured for the following specific WM tracts: left and right dorsal fasciculus gracilis, dorsal fasciculus cuneatus and lateral corticospinal (CST). The values were computed at a single slice centered at the C3 vertebral body as shown in figure (1). The mean, standard deviation and variability of the measurements were then compared between tracts using COV (coefficient of variations) in table (1). An ANOVA test was performed to test the effect of laterality (left versus right) and functionality (motor versus sensory).
Results:
Figure (2) shows WM atlas overlaid onto a b0 image. The white matter tracts automatically identified and selected for further quantitative analysis are illustrated in this figure ( green (1): right CST, green (2): left CST, dark blue (3): right fasciculus cuneatus, dark blue (4): left fasciculus cuneatus, red (5): right fasciculus gracilis, red (6): left fasciculus gracilis, yellow: gray matter, light blue: the unlabeled white matter ). Results of ANOVA on the selected tracts using FA show no effect for laterality (p=0.74) and no effect for functionality of tracts (p= 0.85).
Conclusion:
To the best of our knowledge, this work is the first to create WM atlas of pediatric spinal cord. The pipeline incorporates unique post-processing, followed by template registration and quantification of DTI metrics using atlas-based regions. This automatic method eliminates the need for manual region of interest analysis of various WM tracts and therefore increases accuracy of the measurements. Future work with a larger cohort as well as assessing other DTI indices (Radial Diffusivity (RD) and Axial Diffusivity (AD)) is warranted.
Acknowledgements
No acknowledgement found.References
[1]: De Leener, B., Lévy, S., Dupont, S. M., Fonov, V. S., Stikov, N., Collins, D. L., ... & Cohen-Adad, J. (2017). SCT: Spinal Cord Toolbox, an open-source software for processing spinal cord MRI data. Neuroimage, 145, 24-43.[2]: De Leener, B., Fonov, V. S., Collins, D. L., Callot, V., Stikov, N., & Cohen-Adad, J. (2018). PAM50: unbiased multimodal template of the brainstem and spinal cord aligned with the ICBM152 space. Neuroimage, 165, 170-179.
Figures
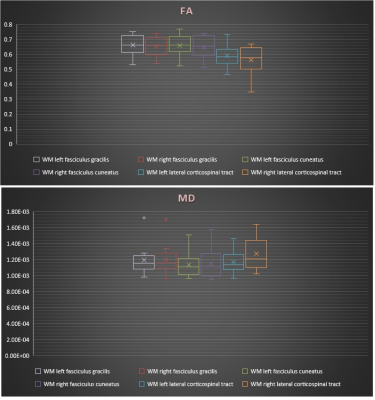
Figure (1): FA and MD for selected WM tracts
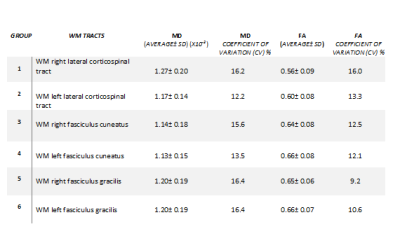
Table (1): Shows the mean, standard deviation and coefficient of variations.

Figure (2): WM atlas overlaid onto a b0 image. Selected WM tracts: green (1): right CST, green (2): left CST, dark blue (3): right fasciculus cuneatus, dark blue (4): left fasciculus cuneatus, red (5): right fasciculus gracilis, red (6): left fasciculus gracilis, yellow: gray matter, light blue: the unlabeled white matter