4571
T2-Weighted Foetal Brain MRI: Utilising Slice-To-Volume Reconstruction to Reduce Repeat Acquisition Due to Fetal Motion1Centre for the Developing Brain, School of Biomedical Engineering and Imaging Sciences, Kings College London, London, United Kingdom
Synopsis
Single-shot turbo spin echo T2 (SSTSE) is a common sequence
used in fetal brain MRI. It is typically acquired in stacks that are positioned
according to fetal brain anatomy, producing images that are radiologically
in-plane; axial, sagittal, and coronal. However, fetal motion can cause images
to be out-of-plane and repeats are often necessary to rectify this. This study
proposes a new, optimised method that acquires stacks in planes set to maternal
anatomy, rather than fetal brain. We then utilise slice-volume reconstruction
to re-register the volume to standard radiological planes.
Introduction
A major challenge for fetal brain MRI is motion. The standard sequence is the T2-weighted single-shot turbo spin echo (ssTSE), which freezes motion in each acquired slice, but there is often change in fetal positon between slices. Interleaved slice acquisition order can reduce slice cross talk, improving robustness [1], but increasing geometrical inconsistency. Conventionally, clinical practice is to “chase the fetus” in order to align with fetal brain anatomy, producing axial, sagittal, or coronal in-plane images (Figure 1). Since fetal pose can change between planning and acquisition, the need to repeat acquisitions to capture the desired planes is common. In addition, focusing on fetal anatomy risks fold over of maternal tissue, leading to either artefacts or need for increased field of view (FoV).MRI reconstruction methods now make it possible to produce coherent 3D volumetric images using 2D stacks. One such approach, Slice-to-Volume Reconstruction (SVR) [2][3][4] uses image registration to align each individual slice into a self-consistent anatomical volume, which is then reconstructed using scattered data interpolation.
SVR does not require slices to be in standard anatomical planes, yet it is common practice to seek to acquire these, resulting in increased examination times if acquisitions are repeated to achieve standard views when fetal pose changes between planning and acquisition. Since SVR integrates slice data regardless of its specific anatomical content, a logical approach could be to abandon the fetal native anatomical focus in favour of a more efficient strategy designed for 3D reconstruction. The proposed optimised method acquires 2D stacks using set planes according to maternal anatomy (Figure 2). This does not follow fetal brain anatomy and will not be in-plane. Once reconstructed, the SVR process re-aligns the 3D volume to axial, sagittal, and coronal fetal planes ready for radiological reporting. This method also enables a smaller FoV as positioning is not dependant on fetal position.
Our aim was to compare the 3D image quality and efficiency of both 2D acquisition methods.
Methods
Imaging was acquired between May 2017 and April 2019, on a Philips 3T Achieva (Best, The Netherlands) using a 32-channel cardiac coil. Only datasets with completed protocol were included. A retrospective analysis was performed on the two approaches and assessed for image quality and efficiency. Forty-three women were referred for clinical fetal brain MRI, and consented for research under the London-West Ethics Committee (HO/707/105). Median (range) gestational age (GA) 28+ 4 (20-40) weeks.Seventy-four women were recruited for fetal brain research MRI, and consented to (Fulham Ethics Committee) Placenta Imaging Project where fetal brain images were acquired as part of the imaging protocol. https://placentaimagingproject.org/project/. Median GA 28+1 (20-40) weeks. The clinical group were imaged using an existing “standard” fetal centric approach (Clinical Protocol), acquiring standard T2ssTSE axial, sagittal, and coronal planes of the fetal brain (Figure 1). Parameters; TR=32445ms, TE=180ms, Acquired Voxel Size=1.25x1.26x2.5mm, Slice Gap=-1.25mm, Sense = 2, Number of Slices = 79 (axial), 84 (sagittal), 98 (coronal), FoV=320x450x106mm (sagittal), 320x450x100mm (axial), 320x450x124mm (coronal), total number of dynamics = 6. Total acquisition time=15.5minutes. Total acquisition time may be longer if acquisitions need repeating due to excess motion or fetus moving out-of-plane.
The research group were imaged using a time optimized maternal centric approach (Optimized Protocol), acquiring non-standard planes positioned according to maternal anatomy (Figure 2). Parameters: TR=26663ms, TE=180ms, Acquired Voxel Size=1.51x1.52x2.5mm, Slice Gap=-1mm, Sense=2, FoV=320X320X121mm(axial), 320x350x121mm (sagittal-oblique), Number of Slices=80, total number of dynamics=6, total acquisition time= 8.58minutes.
Prior to SVR all acquired stacks were assessed for motion, those with severe corruption were excluded from the SVR pipeline. The 3D volume was awarded a quality score using a 4-point scoring system, deducting a point for each of the following: B1 artefact, Blurring, Signal Loss, and/or Gross Artefact (Figure 3). The 2D stacks and the completed SVR volumes were then reported by a consultant neuroradiologist. 2D stacks for both methods are shown alongside the final SVR image. (Figure 4).
The number of T2 stacks repeated outside the normal protocol was calculated and a chi-squared test used to assess for significant differences in the number of repeats performed in each method.
A Wilcoxon Rank Sum test was used to compare SVR image quality scores for both approaches.
Results
There was no significant difference in GA between subject groups (p = 0.86).One SVR failed in the Clinical Protocol, one failed in the Optimised Protocol.
The Clinical Protocol had 100 of 347 acquired stacks out-of-plane due to fetal motion. Out of 43 clinical datasets 22 had repeated acquisitions. Out of 74 optimized datasets 10 had repeated acquisitions.
There was no significant difference in quality score (z = -0.48, p = .631).
Significantly less T2 stacks were repeated using the Optimized Protocol compared to the Clinical Protocol. Pearson Chi-Squared test = 4, df=28.177, p<0.01. (figure 5).
Conclusion
The clinical method of acquisition remains the mainstay for radiological reporting, but fails to deliver in-plane images without potentially extending acquisition time for repeat acquisitions. In addition, slice angulation is dependent on fetal pose, leading to possible aliasing. Our optimized method was able to produce radiographic images of comparable quality, significantly reduce the number of repeats, and reduced acquisition time.Acknowledgements
This work was supported by the NIH Human Placenta Project grant 1U01HD087202-01 (Placenta Imaging Project (PIP), the Wellcome EPSRC Centre for Medical Engineering at Kings College London (WT 203148/Z/16/Z) and by the National Institute for Health Research (NIHR) Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health.References
1. Koberlein G., Hosseinzadeh K., Anthony E.Y. Fetal MR Imaging: Protocols and Anatomy. In: Masselli G. (eds) MRI of Fetal and Maternal Diseases in Pregnancy. Switzerland: Springer International Publishing Switzerland; 2016: p.35-56.
2. Murgasova-Kuklisova. M., Quaghebeur. G., Rutherford. M., & Hajnal. J.V. Reconstruction of fetal brain MRI with intensity matching and complete outlier removal. Medical Image Analysis. 2012; 16(8): 1550-1564. Available from: https://www.sciencedirect.com/science/article/pii/S1361841512000965 [Accessed 1st November 2019].
3. Kainz, B., Steinberger. M., Wein. W., Murgasova-Kuklisova. M., Malamateniou. C., Kerauduren. K., Torsney-Wier. T., Rutherford. M., Aljabar. P., Hajnal. J.V., & Rueckert. D. Fast Volume Reconstruction From Motion Corrupted Stacks of 2D Slices. IEEE Transactions on Medical Imaging. 2015; 34(9): 1901-1913. Available from https://ieeexplore.ieee.org/document/7064742/authors#authors[Accessed 1st November 2019]
4. Rousseau, F., Glenn, O. A., Iordanova, B., Rodriguez-Carranza, C., Vigneron, D. B., Barkovich, J. A., & Studholme, C. Registration-Based Approach for Reconstruction of High-Resolution In Utero Fetal MR Brain Images. Academic Radiology. 2006; 13(9), 1072–1081.
Figures
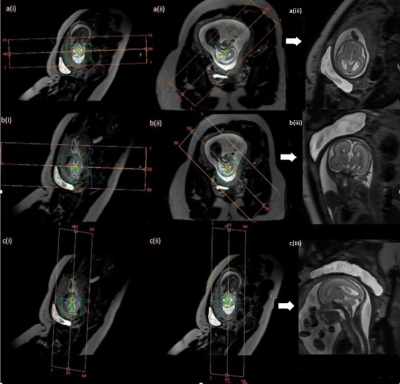

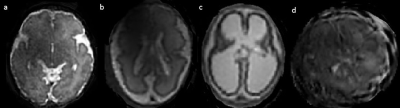
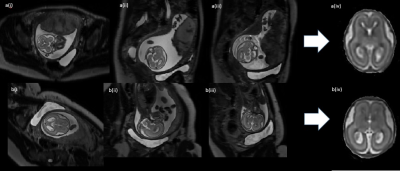
Figure 4: Images from both approaches, with completed SVR (top row) fetal brain (GA=21+0) using Optimized Protocol with SVR: a(i) stack covering fetal brain axial to the maternal pelvis, a(ii) stack covering fetal brain sagittal-oblique to maternal pelvis, a(iii) stack covering fetal brain sagittal oblique to maternal pelvis, a(iv) completed SVR image. (bottom row) Fetal brain (GA=21+2) using Clinical Protocol with SVR: b(i) stack aligned axial to fetal brain, b(ii) stack acquired sagittal to fetal brain, b(iii) stack aligned coronal to fetal brain, b(iv) Completed SVR image.