4530
Topological alterations in structural brain connectivity networks are associated with survival after out-of-hospital cardiac arrest1Department of Neuroscience and Biomedical Engineering, Aalto University School of Science, Espoo, Finland, 2Turku Brain and Mind Center, University of Turku, Turku, Finland, 3Turku PET Centre, University of Turku and the Hospital District of Southwest Finland, Turku, Finland, 4Department of Radiology, Turku University Hospital, University of Turku, Turku, Finland, 5Department of Medical Physics, Turku University Hospital, University of Turku, Turku, Finland, 6Division of Perioperative Services, Intensive Care and Pain Medicine, Turku University Hospital, University of Turku, Turku, Finland, 7Division of Intensive Care Medicine, University of Helsinki and Helsinki University Hospital, Helsinki, Finland, 8Department of Radiology, University of Helsinki and Helsinki University Hospital, Helsinki, Finland, 9Department of Neurology, University of Helsinki and Helsinki University Hospital, Helsinki, Finland, 10Division of Clinical Neurosciences, Turku University Hospital, University of Turku, Turku, Finland
Synopsis
Mortality after out-of-hospital cardiac arrest is high, and there is a substantial need for new biomarkers to improve the identification of patients with poor outcome. Therefore, we investigated structural brain connectivity networks in patients after out-of-hospital cardiac arrest in order to detect differences related to survival. We found decreased global efficiency and strength from MRI scans acquired in a median of 53 hours (IQR 47-64) after OHCA to be related to mortality at 6 months after OHCA. In addition, several regions with decreased strength and local efficiency were found, most significantly in the pallidum, and superior frontal and supramarginal cortices.
Introduction
Mortality after successfully resuscitated out-of-hospital cardiac arrest (OHCA) is high, from 41% to 86%, primarily due to hypoxic-ischemic brain damage1-2. In addition, the survivors have a high risk for a variety of neurological injuries3. Poor outcome cannot be reliably predicted by the multimodal approaches currently in clinical use4-5. Therefore, there is a substantial need for new biomarkers to improve the identification of patients with poor neurological outcome.Diffusion magnetic resonance imaging (dMRI) has enabled the noninvasive investigation of neural tracts and their microstructural properties in vivo6. However, traditional dMRI methods underestimate the extent of ischemic injury during the first three days after OHCA7. Therefore, we used a new method, structural brain connectivity networks, to investigate comatose patients after OHCA. The hypothesis was that the brain networks reconstructed from MRIs acquired in a median of 53 hours (interquartile range 47-64 hours) after OHCA would be different in survivors compared to those who died within 6 months of the OHCA.
Methods
This study is part of the randomized phase II clinical drug trial (Xe-Hypotheca trial; ClinicalTrials.gov identifier: NCT00879892) in which 224 consecutive comatose survivors of witnessed out-of-hospital cardiac arrest from an initial shockable rhythm admitted to the Turku and Helsinki University hospitals between August 2009 and September 2014 were screened for eligibility as described earlier8. A total of 110 patients were enrolled and allocated in a 1:1 ratio to receive either therapeutic hypothermia treatment alone for 24 hours or inhaled xenon in combination with hypothermia for 24 hours. We have previously reported the primary and secondary clinical end points of the Xe-HYPOTHECA trial8-9. Of the 110 patients, 97 underwent magnetic resonance imaging in a median (inter-quartile range) time of 53 hours (47-64) after OHCA and 96 were included in this study.Patients were kept intubated and sedated until MRI was performed regardless of neurological status. The study was approved by the ethics committee of the Hospital District of Southwest Finland and the institutional review boards of the Helsinki University Hospital and the Finnish Medicines Agency. All patients’ next of kin or legal representative gave written informed assent within 4 hours after hospital arrival.
Diffusion MRI data were acquired by using 20 gradient orientations imaged twice with a b‑value of 1000 s/mm2 with a resolution of 2 mm × 2 mm × 3 mm. Sagittal T1‑weighted MRI data were acquired with a resolution of 1 mm × 1 mm × 1 mm.
The DW data were denoised10 and corrected for subject motion11, bias field12, eddy current induced13, and echo planar imaging distortions14. The parcellation of the T1-weighted images was performed in FreeSurfer15 using Desikan-Killiany atlas16 combined with the subcortical gray matter structures segmented with FSL's17 FIRST18, resulting in 84 gray matter regions. Structural brain connectivity networks19-20 were reconstructed in MRtrix21 by combining the parcellation and the streamlines reconstructed with CSD-based tractography22. Anatomically constrained tractography23 was used to improve the validity of the reconstructed streamlines. The number of streamlines connecting a pair of regions was used as the connection weight24, resulting in connectivity matrices of 84 × 84.
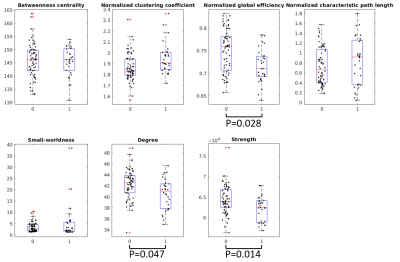
Graph theoretical analysis was used to investigate both global and local properties of the structural brain connectivity networks19-20. In the global analyses, we investigated betweenness centrality25, normalized global efficiency26, normalized characteristic path length27, normalized clustering coefficient28-29, small-worldness27, degree, and strength. Local node-level analyses were performed for local efficiency26 and strength. Normalization was performed by comparing to 100 randomized networks with equal weight, degree, and strength distributions30. Age, gender, and imaging site were used as covariates in all statistical analyses. False discovery rate (FDR) was used to correct for multiple comparisons with a significance threshold of α=0.05.
Results
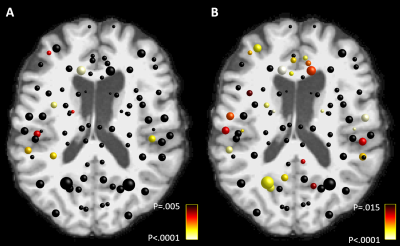
In the global network properties, we found decreased global efficiency, degree, and strength in those who died after OHCA compared to the survivors, as shown in Figure 1.In the local analyses, we found 8 regions with decreased strength, and 25 regions with decreased local efficiency in those who did not survive compared to the survivors, as shown in Figure 2. No regions showed increases in these metrics.
Discussion
In this study, survival was associated with global and local properties of the structural brain connectivity networks in patients after OHCA. Decreased integration and strength of the networks reconstructed based on MRI data acquired in a median of 53 hours (interquartile range 47-64 hours) after OHCA were found in patients who did not survive 6 months after OHCA. In addition, 8 regions with decreased strength and 25 regions with decreased efficiency were associated with mortality. Right hemisphere was more significantly affected. Of these regions, four were significantly different in both strength and local efficiency: right pallidum, superior frontal cortex, supramarginal cortex, and pars orbitalis. Bilateral differences were observed in the pallidum, superior frontal, superior temporal, transverse temporal, supramarginal, and medialorbitofrontal cortices.A limitation of the study is a suboptimal acquisition protocol for CSD due to a low b-value and a low number of gradient orientations31.
Conclusion
We found decreased integration, degree, and strength of the structural brain connectivity networks to be associated with mortality after OHCA. In addition, decreased strength and efficiency of several nodes were associated with higher mortality.Acknowledgements
T.R. received funding from the Emil Aaltonen Foundation, Finland and the Finnish Cultural Foundation, Finland. U.R received funding from Finnish Medical Foundation and Arvo and Lea Ylppö Foundation, Finland.References
1. Graesner J, Lefering R, Koster RW, et al. EuReCa ONE-27 Nations, ONE Europe, ONE Registry A prospective one month analysis of out-of-hospital cardiac arrest outcomes in 27 countries in Europe. Resuscitation. 2016;105:188-195.
2. Nielsen N, Wettersley J, Cronberg T, et al. Targeted Temperature Management at 33 degrees C versus 36 degrees C after Cardiac Arrest. N Engl J Med. 2013;369(23):2197-2206.
3. Dragancea I, Rundgren M, Englund E, et al. The influence of induced hypothermia and delayed prognostication on the mode of death after cardiac arrest. Resuscitation. 2013;84(3):337-342.
4. Nolan JP, Soar J, Cariou A, et al. European Resuscitation Council and European Society of Intensive Care Medicine Guidelines for Post-resuscitation Care 2015 Section 5 of the European Resuscitation Council Guidelines for Resuscitation 2015. Resuscitation. 2015;95:202-222.
5. Sandroni C, D'Arrigo S, Nolan JP. Prognostication after cardiac arrest. Crit Care. 2018;22(1):150.
6. Tournier JD, Mori S, Leemans A. Diffusion tensor imaging and beyond. Magnet Reson Med. 2011;65(6):1532-1556.
7. Wijman CAC, Mlynash M, Caulfield AF, et al. Prognostic Value of Brain Diffusion-Weighted Imaging after Cardiac Arrest. Ann Neurol. 2009;65(4):394-402.
8. Laitio R, Hynninen M, Arola O, et al. Effect of Inhaled Xenon on Cerebral White Matter Damage in Comatose Survivors of Out-of-Hospital Cardiac Arrest A Randomized Clinical Trial. JAMA. 2016;315(11):1120-1128.
9. Arola O, Saraste A, Laitio R, et al. Inhaled Xenon Attenuates Myocardial Damage in Comatose Survivors of Out-of-Hospital Cardiac Arrest: The Xe-Hypotheca Trial. J Am Coll Cardiol. 2017;70(21):2652-2660.
10. Veraart J, Novikov DS, Christiaens D, et al. Denoising of diffusion MRI using random matrix theory. NeuroImage. 2016:142,394-406.
11. Leemans A, Jones DK. The B‐matrix must be rotated when correcting for subject motion in DTI data. Magnet Reson Med. 2009;61(6):1336-1349.
12. Tustison N, Avants B, Cook P, et al. N4ITK: Improved N3 Bias Correction. IEEE Trans Med Imag. 2010;29:1310-1320.
13. Andersson JL, Sotiropoulos SN. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. NeuroImage. 2016;125:1063-1078.
14. Irfanoglu MO, Walker L, Sarlls J, et al. Effects of image distortions originating from susceptibility variations and concomitant fields on diffusion MRI tractography results. NeuroImage. 2012;61(1):275-288.
15. Fischl B. FreeSurfer. NeuroImage. 2012;62(2):774-781.
16. Desikan RS, Ségonne F, Fischl B, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage. 2006;31(3):968-980.
17. Jenkinson M, Beckmann CF, Behrens TE, et al. FSL. NeuroImage. 2012;62(2):782-790.
18. Patenaude B, Smith SM, Kennedy DN, et al. A Bayesian model of shape and appearance for subcortical brain segmentation. NeuroImage. 2011;56(3):907-922.
19. Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci. 2009;10(3):186.
20. Rubinov M, Sporns O. Complex network measures of brain connectivity: uses and interpretations. NeuroImage. 2010;52(3):1059-1069
21. Tournier JD, Smith R, Raffelt D, et al. MRtrix3: A fast, flexible and open software framework for medical image processing and visualisation. NeuroImage. 2019;202:116137.
22. Tournier JD, Calamante F, Connelly A. Robust determination of the fibre orientation distribution in diffusion MRI: non-negativity constrained super-resolved spherical deconvolution. NeuroImage. 2007;35(4):1459-1472.
23. Smith RE, Tournier JD, Calamante F, et al. Anatomically-constrained tractography: improved diffusion MRI streamlines tractography through effective use of anatomical information. NeuroImage. 2012;62(3):1924-1938.
24. Roine T, Jeurissen B, Perrone D, et al. Reproducibility and intercorrelation of graph theoretical measures in structural brain connectivity networks. Med Imag Anal. 2019;52:56-67.
25. Brandes U. A faster algorithm for betweenness centrality. J Math Sociol. 2001;25:163–177.
26. Latora V, Marchiori M. Efficient behavior of small-world networks. Phys Rev Lett. 2001;87:198701.
27. Watts DJ, Strogatz SH. Collective dynamics of “small-world” networks. Nature. 1998;393:440-2.
28. Onnela JP, Saramäki J, Kertész J, et al. Intensity and coherence of motifs in weighted complex networks. Phys Rev E - Stat Nonlinear Soft Matter Phys. 2005;71(6):065103.
29. Saramäki J, Kivelä M, Onnela JP, et al. Generalizations of the clustering coefficient to weighted complex networks. Phys Rev E - Stat Nonlinear Soft Matter Phys. 2007;75(2):027105.
30. Rubinov M, Sporns O. Weight-conserving characterization of complex functional brain networks. NeuroImage. 2011;56:2068–2079.
31. Tournier JD, Calamante F, Connelly A. Determination of the appropriate b value and number of gradient directions for high‐angular‐resolution diffusion‐weighted imaging. NMR Biomed. 2013;26(12):1775-1786.
Figures