4485
Application of Diffusion MRI ODF-Fingerprinting for Neurosurgical Tractography1Center for Biomedical Imaging, Dept. of Radiology, NYU School of Medicinie, New York, NY, United States, 2Center for Advanced Imaging Innovation and Research (CAI2R), NYU School of Medicine, New York, NY, United States, 3Department of Neurosurgery, Perlmutter Cancer Center, Neuroscience Institute, Kimmel Center for Stem Cell Biology, NYU School of Medicinie, New York, NY, United States
Synopsis
The ODF-Fingerprinting method has proven to be successful in improving detection of fiber pairs with small crossing fibers [1]. We evaluated the performance of ODF-Fingerprinting and other algorithms used to detect fiber directions, to determine whether the clinical application contains the same success that was experienced in a healthy control. The performance of these competing algorithms was examined in a healthy control and post-operative scan of a tumor patient. With the success of ODF-Fingerprinting in comparison to other algorithms, we hope that the application of this algorithm will lead to improved neurosurgical precision.
Methodology
Purpose:Diffusion tractography is routinely used to study white matter architecture and brain connectivity $$$\textit{in vivo}$$$ [2,3]. An essential component for successful tractography of neuronal tracts is the correct identification of the tract directionality in each voxel. However, accurate fiber direction estimation is difficult due to the limited angular resolution of the acquisition and intrinsic ODF peak width [4,5] and most methods fail to detect fibers crossing at angles smaller than 30°-40° [3,5]. Research has indicated that the ODF Fingerprinting methodology can improve the detection of these fiber crossings [1,7,8] by drawing a comparison between the measured ODF and the elements in a pre-generated library of ODF-fingerprints [9,10]. We applied this methodology in a clinical post-operative case to determine the feasibility of ODF-fingerprinting in comparison to several other fiber directionality identification algorithms $$$\textit{in vivo}$$$ .
Methods:
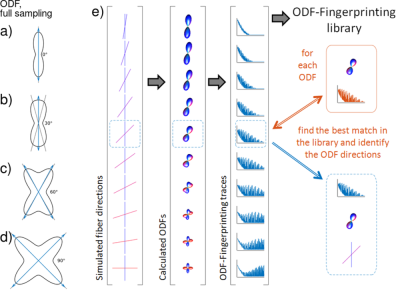
ODF-Fingerprinting: For ODF-Fingerprinting, a library $$$L_{ODF}$$$ was generated by utilizing diffusion weighted signals as a sum of diffusion tensors (FA$$$\,=\,$$$[0.3,1], ADC$$$\,=\,0.9\,\mathrm{mm^2/s}$$$, 10% water component, assumption of cylindrical fibers $$$\lambda_2=\lambda_3$$$) on a Radial Diffusion Spectrum Imaging grid (RDSI, 236 q-space samples, four shells, bmax=$$$4000\,\mathrm{s/mm^2}$$$) and reconstructing ODFs, as demonstrated in Figure 1 [11]. Matching of a measured $$$ODF_m$$$ with the elements of library $$$L_{ODF}$$$ was assessed by maximizing the dot-product [9,10] with an added penalty term for the noise estimate $$$\sigma_n$$$ of the input diffusion data: $$$\mathrm{max}\left(\log(L_{ODF}\,.\,ODF_m) – n_{par}/{4n} \sigma_n\right)$$$ ($$$n_{par} = 1 + 5N_{fib}$$$ a measure of library element complexity and $$$n$$$ the number of elements in $$$ODF_m$$$) [8]. $$$\sigma_n$$$ is estimated using a linear mean square error estimator for the variance of the noise of diffusion weighted signals [12]. The size of the fingerprint-library is reduced by rotating the maximum value of ODF-traces to the Z-axis before matching. The ODF fingerprinting method is compared with local maximum search (DSIStudio), Newton search along a set of specified directions (MRtrix3, $$$\mathrm{sh2peaks}$$$) and constrained spherical deconvolution (CSD, MRtrix3, $$$\mathrm{dwi2fod\,\,\,msmt\_csd}$$$ and $$$\mathrm{sh2peaks}$$$).
Data: An $$$\textit{in vivo}$$$ Radial DSI [11] dataset was acquired on a 3T clinical scanner (3T Siemens Prisma; 20ch head coil; b$$$_{max}\,=\,4000\,\mathrm{s/mm^2}$$$; 236 q-space volumes; TR/TE=2600/119; 45slices; $$$220\,\mathrm{mm}$$$ FoV; $$$3\,\mathrm{mm}$$$ isotropic resolution; PF5/8; male volunteer 60y/o). The images are denoised [14] and corrected for susceptibility, eddy currents and subject motion using eddy from the FSL Library [13]. RDSI reconstructions, incorporating variable sample density correction, were performed offline using custom-made software (Matlab, Mathworks). Display with Matlab and DSIStudio [Yeh2010].
Tractography: Corticospinal (CST), Arcuate Fasciculus (AF) and Optic Radiation (OR) tracts were generated using DSIStudio [Yeh2010] by seeding from the precentral lobe, the operculum and the visual cortex respectively. Fiber tracts are generated with a tracking algorithm (Yeh et al., 2013, implemented in DSIStudio (Yeh et al., 2010), parameters as suggested in (Fernandez-Miranda et al., 2012)) in both simulated and in vivo datasets. Tracts originate from random seeding points uniformly distributed in user-defined seeding regions, propagate along the most prominent fiber direction with a step size of 2 mm, are smoothed with 20% of the previous direction and halt when the turning angle > 60∘ or QA < 0.25. Resulting fiber tracts shorter than 20mm and longer than 400mm are discarded until a predetermined number of fiber tracts (1 10$$$^4$$$) is created or a maximum number of seed points is reached (1 10$$$^6$$$).
Results and Discussion
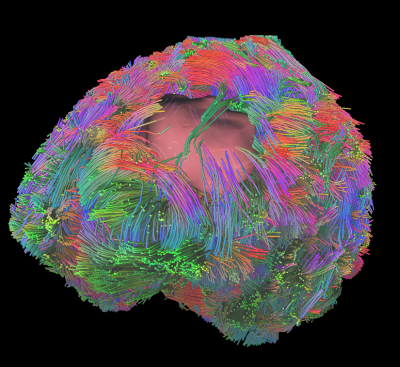
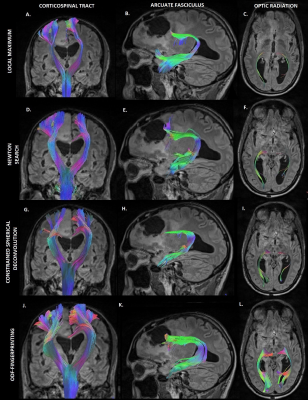
Figure 2 demonstrates the overall fiber tractography of the brain with a tumor embedded in the frontal lobe. The figure demonstrates how the fibers move around the tumor region, which aids in neurosurgical planning. The tracts analyzed by utilizing the ODF-Fingerprinting methodology demonstrate a fuller fanning of fiber groupings. The ODF-Fingerprinting also demonstrated improved fiber tractography in the various regions of interest (Corticospinal, Arcuate Fasciculus, and Optic Radiation) as shown in Figure 3. The figure compares the 3 other tractography methods to the ODF-Fingerprinting, and shows a fuller fanning in the regions of interest with ODF-Fingerprinting.Conclusion
ODF-Fingerprinting has been shown to improve detection of fiber pairs with small crossing angles in simulations and qualitative evaluations of $$$\textit{in vivo}$$$ data. Here we show the application of ODF-FP $$$\textit{in vivo}$$$ in a clinical setting. The improved fiber identification by ODF-FP will aid neurosurgeons in pre-operative planning by generating improved renderings of brain connectivity. Future work will focus on examining additional clinical patients and on verifying the method in fiber phantoms.Acknowledgements
This project is supported in part by PHS grants R01-CA111996, R01-NS082436 and R01-MH00380. DGP is supported by NIH/NINDS R01-NS102665, New York State DOH01-STEM5-2016-00221 and NIH/NIAID R21-AI130618. This work was performed under the rubric of the Center for Advanced Imaging Innovation and Research (CAI2R, https://www.cai2r.net), a NIBIB Biomedical Technology Resource Center (NIH P41EB017183).References
[1] Baete, S. (2019), ‘Fingerprinting Orientation Distribution Functions in diffusion MRI detects smaller angle crossing’, Neuroimage, vol. 198, pp. 231-41.
[2] Wedeen, V.J. (2012), ‘The geometric structure of the brain fiber pathways’, Science, vol. 335, pp. 1628-34.
[3] Tuch, D.S. (2004), ‘Q-ball imaging’, Magn. Reson. Med., vol. 52, pp. 1358-72.
[4] Jeurissen, B. (2013), ‘Investigating the Prevalence of Complex Fiber Configurations in White Matter Tissue with Diffusion Magnetic Resonance Imaging’, Human Brain Mapping, vol. 34, pp. 2747-66.
[5] Barnett, A. (2009), ‘Theory of Q-ball Imaging Redux: Implications for Fiber Tracking’, Magn. Reson. Med., vol 62, pp 910-923
[6] Tournier, J.-D. (2008), ‘Resolving crossing fiberes using constrained spherical deconvolution: Validation using diffusion-weighted imaging phantom data’, NeuroImage, vol 42., pp. 617-25.
[7] Baete, S.H. (2017), ‘Fingerprinting Orientation Diffusion Functions in Diffusion MRI detects smaller crossing angles’, Human Brain Mapping, vol. 23, pp. 1741.
[8] Baete, S.H. (2018), ‘Improving Fingerprint maching for ODFs in Diffusion MRI by using a fiber complexity penalty’, Human Brain Mapping, vol. 24, pp. 2317.
[9] Ma, D. (2013), ‘Magnetic Resonance Fingerprinting’, Nature, vol 495, pp. 187.
[10] Cloos, M. (2016), ‘Multiparametric imaging with heterogeneous radiofrequency fields’, Nature communications, pp. 12445.
[11] Baete, S. (2016), ‘Radial q-Space Sampling for DSI’, Magn. Reson. Med. vol 76, pp. 769-780.
[12] Aja-Fernandez, S., Alberola-Lopez, C., Westin, C. F., 2008. Noise and signal estimation in magnitude mri and rician distributed images: a lmmse approach. IEEE Trans Image Process 17 (8), 1383-98.
[13] Yeh, F.C., 2010. Generalized q-sampling imaging. IEEE Trans Med Imaging 29(9), 1626-35.
Figures