4475
Clinical Recovery of intracellular volume fraction and fiberODF for a patient with asymptomatic temporal-occipital lesion using Deep Learning1Learning Research and Development Center, University of Pittsburgh, Pittsburgh, PA, United States, 2Electrical Engineering & Computer Science, Vanderbilt University, Nashville, TN, United States, 3Department of Neurosurgery, Stanford University, Palo Alto, CA, United States, 4Radiology, Vanderbilt University Medical Center, Nashville, TN, United States, 5Biomedical Engineering, Vanderbilt University, Nashville, TN, United States
Synopsis
Diffusion-weighted magnetic resonance imaging (DW-MRI) offers a unique insight on microarchitecture of the in-vivo human brain. Multiple well-known reconstruction methods that model geometrical and micro-structural properties of the tissue such as multi-tissue constrained spherical deconvolution (MT-CSD) and spherical mean technique (SMT) rely on high quality acquisitions (more than 2 shells and 45 gradient directions) which is a constraint. We propose recovery of fiber-ODFs, compartment diffusivities and volume-fractions using a two-stage deep learning framework by training on human-connectome-project dataset. The proposed approach can predict fiber-ODFs using single shell DW-MRI on a tumor patient and assess the diseased region of interest.
Introduction
Diffusion-weighted magnetic resonance imaging (DW-MRI) is a unique way to map geometrical properties of brain tissues; for example, estimating fiber ODFs and using fiber tractography. Additionally, DW-MRI is also utilized to characterize the micro-structure of human brain using various popular methods like CHARMED, NODDI and SMT. These methods are constrained theoretically on high quality acquisitions (more than 2 shells and 45 gradient directions). High b-values and high angular directions diffusion data are constrained by the limitations of hardware and scanning time in clinical setting. The long scan time of the methods limit their use in time limited billable clinical scans and reduce usage in surgical planning and in diagnosing lesions.We propose recovery of fiber-ODFs, compartment diffusivities and volume-fractions using a two-stage deep learning framework by training on human connectome project datasets. The trained model depicts applicability on a clinical subject with neurological disorder.
Methods
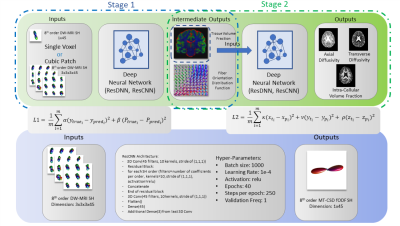
We utilize 15 subjects from Human Connectome Project for training and validation of the deep learning model [2]. The data was split into three pools of training: 5, validation: 2 & testing: 8. The HCP standard acquisition consists of 90 gradient directions for three b-values of 1000, 2000 & 3000 s/mm2. Multi-tissue CSD [4] derived FOD (geometric tissue structure) and volume fraction (intra-tissue property) ground truth were created using all directions per subject. Spherical mean technique [5] ground truth of ICVF, Axial diffusivity and radial diffusivity was generated using all data as well. MT-CSD & SMT ground truth will serve as the outputs of the deep learning approach. Spherical harmonics of 8th order were utilized for representation of a single shell data of HCP [3]. To ensure clinical usage only the diffusivity shell of 1000 s/mm2 was utilized with 90 gradient directions.A 56 year old male presented with asymptommatic, right sided temporo-occipital lesion. He underwent a brain MRI using a General Electric 1.5T scanner as part of a routine pre-surgical staging workup. Scan revealed a mass consistent with a gliomatous neoplasm. Protocol used included b=1000 s/mm2 30 gradient directions, 1x1x2 mm3 voxel size, with 80 axial slices.
We propose to utilize a two-stage deep learning framework for predictions of MT-CSD FOD and SMT metrics (Fig 1.). For the first stage we utilize a prior deep learning proposed approach (ResCNN) for predicting MT-CSD FOD and volume fraction estimates based on cubic patch inputs of DW-MRI SH coefficients [1]. The MT-CSD FOD and volume fraction ground truth of HCP serve as the outputs of patch based deep learning approach. L1 depicts the loss function in Fig 1 and other deep learning hyper-parameters. To derive the SMT based metrics from the single-shell DW-MRI we utilize a secondary network following a similar architecture as in [1]. The inputs for the secondary network are the volume fraction coefficients predicted from the ResCNN and the ground truth of SMT is utilized. We use a per metric mean squared error loss function for the secondary network L2.
Results
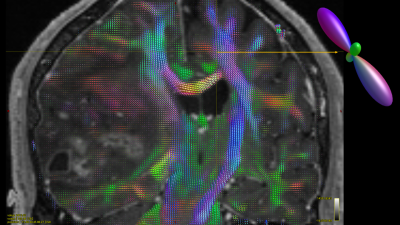
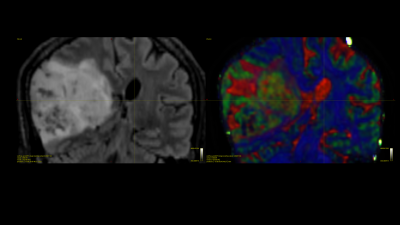
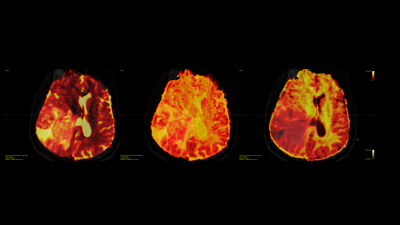
Prior results on the 8 testing subjects indicate high quantitative correlation 0.77 for stage 1 and low error for stage 2 relatively 7-10% error. The estimated deep learning parameters can be successfully used to predict fiber ODFs maps for tumor patients. Fig 2 shows the fODF in cortico-spinal tracts and corpus callosum regions which are consistent with neuroanatomy. Predicted tissue volume fraction is consistent with the FLAIR’s tissue contrast in normal tissue and exhibit differentiation in tissue type for tumor regions (Fig 3). Predicted radial diffusivity (SMT metric) indicates differences in tissue types within the tumor regions (Fig 4). ICVF around the tumor regions shows that white matter tracts are pushed. Corticospinal tract is covered by edema as shown in FLAIR image and can be uncovered using these maps (Fig 5).Discussion
The proposed deep learning framework can learn non-linear mapping of raw single shell diffusion data to complex fiber-ODF for geometrical mapping and highly nonlinear micro-structural maps. This framework can be very useful for clinical acquisitions for pre-surgical mapping.In this particular case, cortico-spinal tract (CST) is affected by the tumor and surrounded edema. CST is shifted and partially pass through the edema region. CST is unaffected by edema because current model separate different tissue compartments within voxel.
Second stage of the framework is used to predicted micro-structural tissue contrast. SMT based diffusivities and ICVF are estimated for each voxel. These maps show different tissue types within the tumor affected regions, which suggests that this framework is capable to provide different tissue contrast (cellularity, edema etc).
Conclusion
We have shown that a deep learning framework that can be used to learn geometrical and microstructural maps between routine diffusion scan in clinics and high spatial and angular resolution DW-MRI. These maps provide key clinical information about the affected tissue and help neurosurgeons to plan surgery. We have shown fiber tracts estimated using the predicted maps on a tumor patient and tissue properties around within the tumor region.Acknowledgements
No acknowledgement found.References
- Vishwesh Nath, Sudhir K. Pathak, Kurt G. Schilling, Walter Schneider, Bennett A. Landman "Deep Learning Estimation of Multi-Tissue Constrained Spherical Deconvolution with Limited Single Shell DW-MRI", SPIE 2020, Medical Imaging, Houston, TX, USA
- Van Essen, D.C., et al., The WU-Minn human connectome project: an overview. 2013. 80: p. 62-79.
- Descoteaux, M., et al., Apparent diffusion coefficients from high angular resolution diffusion imaging: Estimation and applications. Magnetic Resonance in Medicine, 2006. 56(2): p. 395-410.
- Jeurissen, B; Tournier, J-D; Dhollander, T; Connelly, A & Sijbers, J. Multi-tissue constrained spherical deconvolution for improved analysis of multi-shell diffusion MRI data NeuroImage, 2014, 103, 411-426
- Kaden E, Kelm ND, Carson RP, Does MD, and Alexander DC: Multi-compartment microscopic diffusion imaging. NeuroImage, vol. 139, pp. 346–359, 2016
Figures