4472
Redundancy of arousal brainstem structural connectivity pathways in humans by 7 Tesla HARDI1Department of Radiology, Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital and Harvard Medical School, Boston, MA, United States
Synopsis
Arousal plays a crucial role in wakefulness/sleep, autonomic function, affect and attention. Recent meta-analysis of rat neuroanatomical macroconnectome based on pathway-tracing studies showed redundancy of arousal mechanism through the presence of multiple connectivity pathways and of high interconnectivity among arousal brainstem nuclei. Using 7 Tesla HARDI MRI and a recently developed brainstem nuclei atlas, we build the structural connectome of brainstem arousal nuclei in living humans. In line with rat studies, this connectome showed high redundancy through the presence of multiple interconnected pathways. This connectome might be used to better understand arousal and its impairment.
Introduction
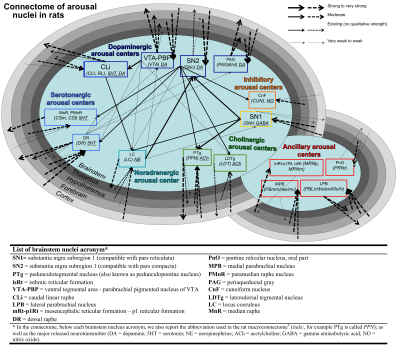
Arousal plays a crucial role in a wide variety of phenomena including wakefulness/sleep, autonomic function, affect and attention1. In the past 70 years1-2 several brainstem nuclei have been implicated in the maintenance of arousal based on animal stimulation/lesions studies and on their ability to modulate the cortex either through direct connectivity pathways to the cortex or through the thalamus, hypothalamus and basal forebrain. Recent meta-data analysis3 of rat neuroanatomical macroconnectome based on pathway-tracing studies4 showed properties of redundancy of arousal mechanism through the presence of multiple connectivity pathways stemming from arousal brainstem nuclei and of high interconnectivity among them (Figure 1). Interestingly, a redundant arousal network might provide a neural and conceptual framework to understand/optimize spontaneous/drug-induced compensatory mechanisms in disorders associated to impairments in arousal (e.g. disorders of consciousness and sleep disorders). Nevertheless, a diagram of brainstem arousal structural connectivity pathways and evaluation of their redundancy are missing in living humans due to limitations in conventional diffusion MRI and the lack of localization of these tiny brainstem nuclei in vivo.Purpose
To investigate the structural connectivity of arousal brainstem nuclei in humans by the use of high-spatial and high-angular resolution diffusion imaging (HARDI) at 7 Tesla, as well as a recently developed in-vivo probabilistic structural atlas of arousal brainstem nuclei5-6 in stereotactic (Montreal-Neurological-Institute —MNI) space.Methods
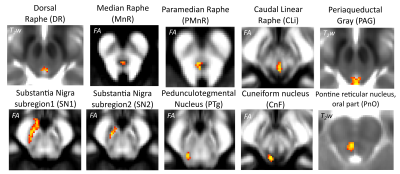
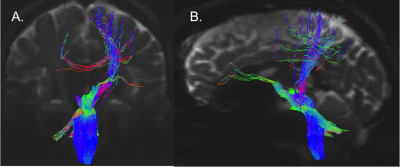
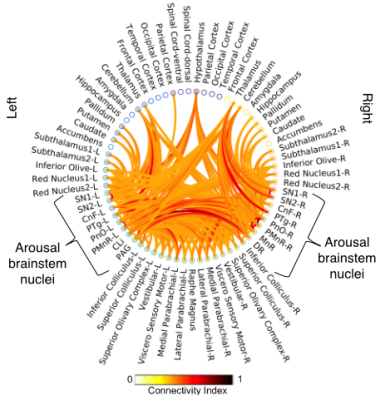
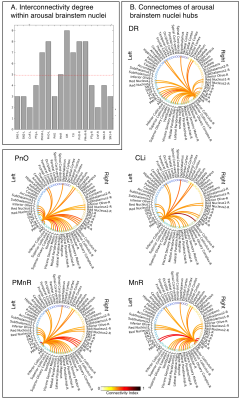
Data acquisition: Twelve healthy subjects (6m/6f, age 29±3 ) underwent MRI under IRB-approval using a 7 Tesla whole-body scanner. T1-weighted multi-echo MEMPRAGE image: with parameters repetition-time/echo-times/inversion time/flip-angle/FOV/bandwidth/GRAPPA-factor: 2.51s/1.6, 3.5, 5.3, 7.2 ms/1.5 s/7°/256×256×176 mm3/“651 Hz/pixel”/2, acquisition-time: 6′34′′. HARDI: common single-shot 2D spin-echo-EPI (using a prototype sequence which supports unipolar diffusion-encoding) with parameters n. slices/echo-time/repetition-time/phase-encoding direction/bandwidth/partial-Fourier/n. diffusion-directions/b-value: 82 /66.8 ms/7.4 s/“anterior/posterior”/“1456 Hz/pixel”/“6/8”/60/2500 s/mm2, seven interspersed “b0” images (T2-weighted, non-diffusion-weighted, b-value = 0 s/mm2), acquisition-time: 8′53′′. To perform distortion-correction we also acquired seven “b0” images with opposite phase-encoding direction.Data analysis: a) Preprocessing: We computed the root-mean-squareMEMPRAGEimage across echo-times, rotated it to standard-orientation (“RPI”), cropped the most inferior slices containing the spinal-cord (in order to aid its coregistration to MNI-space) and bias-field corrected it (SPM8); we then parcellated the resulting image with Freesurfer7. HARDIs were rotated to standard-orientation, de-noised8, motion and distortion-corrected (FSL, topup/eddy). We then computed the diffusion tensor-invariants (e.g. fractional-anisotropy, FA) and S0 (T2-weighted) image from the preprocessed HARDIs (FSL, dtifit). To map the Freesurfer parcellation to native HARDI-space, we computed an affine boundary-based transformation (FSL, FLIRT-BBR) between the preprocessed MEMPRAGE and single-subject S0 images. To map the brainstem nuclei atlas to native HARDI-space,we computed the bivariate high-dimensional diffeomorphic transformations (ANTs) between IIT-MNI FA/S0 templates9 and single-subject FA/S0 images using an intermediate group-based bivariate optimal-template. b) Definition of seed and target regions for HARDI-based connectivity analysis: As seed regions, we used the structural probabilistic atlas labels5, 6 of ten arousal brainstem nuclei (Figure 2) mapped from IIT-MNI-space to native-space (using the coregistration transformations explained above). As target regions, we used the probabilistic atlas labels of 41-brainstem nuclei5-6, 10-11, as well as the 82-cortical/subcortical bilateral regions obtained in each subject from the MEMPRAGE Freesurfer-parcellation (mapped to native space as explained above). For display purposes, we grouped cortical parcellations within each cerebral lobe (frontal/parietal/temporal/occipital). c) Single-subject HARDI-based connectivity analysis: We performed probabilistic tractography using MRtrix3-iFOD2 algorithm based on constrained spherical deconvolution, with a 120 degree maximum angle between successive steps and a 1 mm minimum streamline-length (Figure 3). We propagated 100,000 streamlines from each seed-mask, and computed a “structural-connectivity-index” (range: [0 1]) for each pair of seed-target masks (= fraction of streamlines propagated from the seed reaching the target mask). d) Group HARDI-based connectivity analysis: We averaged across subjects the structural-connectivity-index of brainstem nuclei with target-regions to yield a group structural connectome of these nuclei. We displayed this connectome using a 2D circular diagram12. e) Graph-theory-based analysis of redundancy: To evaluate the number of parallel arousal pathways, we counted the number of arousal brainstem nuclei having connectivity to the thalamus, hypothalamus or cortex. To evaluate the interconnectivity of arousal pathways, we measured the arousal brainstem nuclei connectivity-degree (i.e. number of links) among them. Hubs were defined as nuclei that displayed the highest –and above the average- connectivity-degree.
Results and Discussion
All the investigated brainstem arousal nuclei displayed structural connectivity to the thalamus, hypothalamus or cortex and among themselves (Figure 4). This demonstrates the high redundancy of arousal mechanism in humans through the presence of parallel connectivity pathways originating from arousal brainstem nuclei and the high-interconnectivity of these nuclei, in line with rodents studies3-4. The arousal brainstem nuclei inter-connectivity degree is shown in Figure 5A. Graph-theory-based analysis showed that five arousal brainstem nuclei were network hubs: oral-pontine-reticular nucleus (bilateral), paramedian raphe (bilateral), median raphe, dorsal raphe and caudallinear raphe. The connectome of these individual nuclei is shown in Figure 5B.Conclusions
We built an in-vivo human 7 Tesla HARDI-MRI based diagram of structural connectivity pathways of ten arousal brainstem nuclei in living humans showing high redundancy of connectivity, in line with animals studies3-4. This might be useful to understand/diagnose/treat disorders associated to impaired arousal.Acknowledgements
MGH-Claflin-Distinguished-Scholar; Harvard-Mind-Brain-Behavior-Faculty-Award; NIH-NIBIB-K01EB019474; NIH-NIDCD-R21-DC015888; Dr. Thorsten Feiweier for providing the diffusion sequence used in this study.References
1. Satpute A, Kragel PA, Feldman Barrett L, et al., Deconstructing arousal into wakeful, autonomic and affective varieties. Neurosci Lett. 2019; 6;693:19-28.
2. Moruzzi G, Magoun HW. Brain stem reticular formation and activation of the EEG, Electroencephalogr. Clin. Neurophysiol. 1949; 1: 455-473.
3. Bianciardi M,Izzy S, Rosen B, et al.,Resilience of Subcortical Arousal Mechanisms in Patients Recovering from Traumatic Coma. Submitted.
4. Bota M, Dong HW, Swanson LW. Combining collation and annotation efforts toward completion of the rat and mouse connectomes in BAMS. Frontiers in Neuroinformatics2012; 6:2.
5. Bianciardi M, Toschi N, Edlow BE, et al., Toward an in vivo neuroimaging template of human brainstem nuclei of the ascending arousal, autonomic, and motor systems. Brain Connect. 2015; 5: 597-607.
6. Bianciardi M, Strong C, Toschi N, et al., A probabilistic template of human mesopontine tegmental nuclei from in vivo 7T MRI. Neuroimage. 2018; 170: 222-230.
7. Desikan RS, Segonne F, Fischl B, et al., An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006; 31: 968-980.
8. Manjon JV, Coupé P, Concha L, et al., Diffusion Weighted Image Denoising Using Overcomplete Local PCA. PLoS ONE. 2013; 8(9): e73021.
9. Varentsova A, Zhang S, Arfanakis K., Development of a high angular resolution diffusion imaging human brain template. Neuroimage. 2014; 91: 177-186.
10. García-Gomar MG, Strong C, Toschi N, et al., In vivo probabilistic structural atlas inf the inferior and superior colliculi, medial and lateral geniculate nuclei and superior olivary complex in humans based on 7 Tesla MRI.Front. Neurosci. 2019; 13(764): 1-15.
11. Singh K, Indovina I, Augustinack JC, et al., Probabilistic atlas of the lateral parabrachial nucleus, medial parabrachial nucleus, vestibular nuclei complex and medullary viscero-sensory-motor nuclei complex in living humans form 7 Tesla MRI. Biorxiv. 2019. (https://www.biorxiv.org/content/10.1101/814228v1)
12. Irimia A, Chambers MC, Torgerson CM, et al., Circular representation of human cortical networks for subject and population-level connectomic visualization. Neuroimage. 2012; 60: 1340-1351.
Figures