4412
Impact of gradient non-linearities on B-tensor diffusion encoding1Neuropsychology, Max Planck Institute for Human Cognitive and Brain Sciences, Leipzig, Germany, 2CUBRIC, School of Physics, Cardiff University, Cardiff, United Kingdom
Synopsis
We investigate the effect of gradient non-linearities (GNL) on free gradient waveform used for B-tensor diffusion encoding. We show the magnitude of the GNL-bias for strong gradients of $$$300 m\text{T}/\text{m}$$$. We derive a closed-form formula of the voxelwise B-tensor under GNL, independent of the choice of gradient waveform used to encode the B-tensor.
Introduction
Gradient non-linearities (GNL) in MRI systems induces spatially-varying deviations from the desired diffusion gradient waveforms. While the strength of GNL is proportional to gradient strength1,2,3, clinical systems can still be significantly impacted1,4,5,6. GNL can greatly affect diffusion model fitting and requires voxelwise modulation1,7. This approach is well suited for models that don't rely on particular types of sampling schemes such as constant b-value shells, as they can directly incorporate the modified scheme in their fitting procedure.B-tensor encoding is a recent diffusion imaging framework opening new ways to probe the tissues microstructure8,9. We can perform B-tensor encoding efficiently using complex time-varying gradient waveforms8,10. In this work, we derive the B-tensor shape under arbitrary GNL-distortions. This also proves the independence from the choice of gradient waveform used to produce the B-tensor. Finally, we showcase the effect of GNL from a human MRI system with $$$\text{G}_{\text{max}}=300\,m\text{T}/\text{m}$$$ in simulated tissue.
Theory
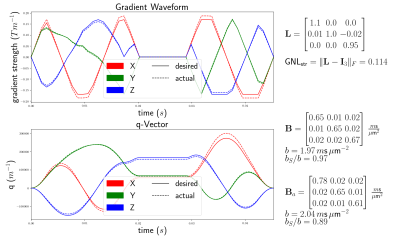
The voxelwise gradient coil tensors $$$\left(\mathbf{L}\right)$$$ are computed from the spatial partial derivative of the effective gradient field1. This field is dependent on the gradient coil geometry and is typically obtained from the vendor. We call "desired" the quantities (gradients, B-tensor, etc) related to the originally intended gradient waveform and "actual" those same quantities under the effect of GNL. We show that the actual B-tensors $$$\left(\mathbf{B}_a\right)$$$ are independent from the desired gradient waveform $$$\left(\vec{G}(t)\right)$$$ used to produce the desired B-tensors $$$\left(\mathbf{B}\right)$$$.From the desired gradient waveform $$$\vec{G}(t)$$$, we define the q-vector as $$$\vec{q}(t) = \gamma \int_0^t \vec{G}(t') \, \text{d}t'$$$. The B-tensor is computed as $$$\mathbf{B} = \int_0^{t_{tot}} \vec{q}(t) \otimes \vec{q}(t) \, \text{d}t$$$ i.e. $$$B_{ij} = \int_0^{t_{tot}} q_i(t)q_j(t) \, \text{d}t$$$ for $$${i,j \in \{x,y,z\}}$$$. The actual gradient waveform under GNL at spatial position $$$\vec{r}$$$ is $$$\vec{G_a}(t) = \mathbf{L}(\vec{r}) \cdot \vec{G}(t)$$$ (fig 1), and the actual q-vector is $$$\vec{q_a}(t) = \mathbf{L} \cdot \vec{q}(t)$$$. Finally, the GNL-distorted actual B-tensor is given by $$$\mathbf{B}_a = [(B_a)_{ij}]$$$ for $$$i,j \in \{x,y,z\}$$$$$\begin{align*}(B_a)_{ij} &= L_{ix} L_{jx} B_{xx} + (L_{ix} L_{jy} + L_{iy} L_{jx}) B_{xy} + (L_{ix} L_{jz} + L_{iz} L_{jx}) B_{xz}\\ & \qquad + L_{iy} L_{jy} B_{yy} + (L_{iy} L_{jz} + L_{iz} L_{jy}) B_{yz} + L_{iz} L_{jz} B_{zz}\end{align*}$$
Experiments and Results
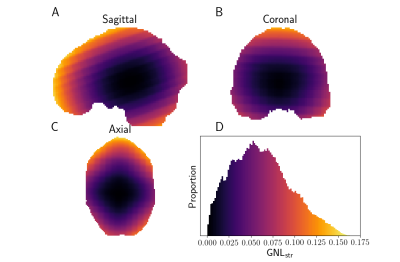
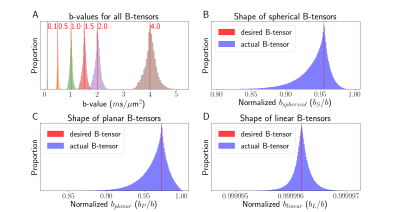
We computed the GNL tensors over a typical brain mask for a Siemens Connectom MRI system $$$\left(\text{G}_{\text{max}}=300 \,m\text{T}/\text{m}\right)$$$. We define the "GNL distortion strength" as $$$\text{GNL}_{\text{str}} = \|\mathbf{L} - \mathbf{I}_3 \|_F$$$. Figure 2 shows the spatial distribution of $$$\text{GNL}_{\text{str}}$$$, which is increasing with the distance from the isocenter of the gradient coil.To showcase the effect of GNL on the desired B-tensor sampling scheme, we generated Maxwell-compensated waveforms10,11 consisting of a total of 384 linear, planar and spherical B-tensors, spanning b-values from 0 to 4 $$$m\text{s}/\mu\text{m}^2$$$ and with main orientation uniformly distributed on the half-sphere. The waveforms were distorted with GNLs tensors within a brain mask, changing their actual b-value and B-tensor shape (fig 3). We use the b-value-normalized tensor-shape metrics8 ($$$b_S/b$$$, $$$b_P/b$$$, $$$b_L/b$$$) defined from the diagonalized B-tensor (eigenvalues $$$b_{1} \le b_{2} \le b_{3}$$$, $$$b = \text{tr}(\mathbf{B}) = b_{1} + b_{2} + b_{3} = b_{S} + b_{P} + b_{L})$$$;
$$\begin{bmatrix}b_{1} & 0 & 0\\0 & b_{2} & 0\\0 & 0 & b_{3}\end{bmatrix}=\frac{b_S}{3}\begin{bmatrix}1 & 0 & 0\\0 & 1 & 0\\0 & 0 & 1\end{bmatrix}+\frac{b_P}{2}\begin{bmatrix}0 & 0 & 0\\0 & 1 & 0\\0 & 0 & 1\end{bmatrix}+b_L\begin{bmatrix}0 & 0 & 0\\0 & 0 & 0\\0 & 0 & 1\end{bmatrix}$$
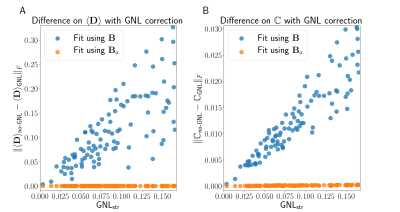
To assess the effect of GNL on estimated diffusion parameters, we simulated a voxel representing fanning white matter pathways. We generated a distribution of diffusion tensors with diffusivities (1.5, 0.3, 0.3) $$$\mu\text{m}^2/m\text{s}$$$ and orientations sampled uniformely in a cone of 30$$$^{\circ}$$$ aperture, centered at direction $$$\left(\frac{1}{\sqrt{3}},\frac{1}{\sqrt{3}},\frac{1}{\sqrt{3}}\right)$$$. We simulated the signal for a uniformly sampled subset of GNL tensors across the entire brain volume. We used the QTI approach9 where the signal is represented by its truncated cumulant expansion $$$\text{S}(\mathbf{B}) \approx \text{S}_0 \exp(-<\mathbf{B}, \langle\mathbf{D}\rangle > + \frac{1}{2} <\mathbf{B} \otimes \mathbf{B} , \mathbb{C}>).$$$ We fit the ensemble-averaged diffusion tensor $$$\left(\langle\mathbf{D}\rangle\right)$$$ and the diffusion tensors covariance matrix $$$\left(\mathbb{C}\right)$$$ using the desired B-tensor $$$\left(\mathbf{B}\right)$$$ and the actual B-tensor $$$\left(\mathbf{B}_a\right)$$$. We report the Frobenius norm of the difference between estimated parameters $$$\left(\langle\mathbf{D}\rangle \text{ and } \mathbb{C}\right)$$$ with $$$\mathbf{B}$$$-fit and with $$$\mathbf{B}_a$$$-fit as an error metric (fig 4). The error produced from ignoring GNL is several orders of magnitude higher than the baseline error and strongly correlated with the $$$\text{GNL}_{\text{str}}$$$ for both $$$\langle\mathbf{D}\rangle$$$ and $$$\mathbb{C}$$$.
Conclusion
GNL have a significant impact on the diffusion-signal and estimated diffusion parameters, especially for strong gradient systems such as the Connectom MRI system. We have derived the closed-form formula of the voxelwise GNL-modified B-tensor. Any diffusion model not relying on exact b-shell or b-tensor shape can incorporate the correction directly into the model fitting procedure.Acknowledgements
MP is supported by a scholarship (PDF-502732-2017) from the Natural Sciences and Engineering Research Council of Canada (NSERC).
CMWT is supported by a Rubicon grant (680-50-1527) from the Netherlands Organisation for Scientific Research (NWO) and a Sir Henry Wellcome Fellowship (215944/Z/19/Z).
CE is supported by the SPP2041 program "Computational Connectomics" of the German Research Foundation (DFG).
References
[1] R. Bammer, M. Markl, A. Barnett, B. Acar, M. Alley, N. Pelc, G. Glover, and M. Moseley, “Analysis and generalized correction of the effect of spatial gradient field distortions in diffusion-weighted imaging”, Magnetic Resonance in Medicine, vol. 50, pp. 560–9, 2003.
[2] S. N. Sotiropoulos, S. Jbabdi, J. Xu, J. L. Andersson, S. Moeller, E. J. Auerbach, M. F. Glasser, M. Hernandez, G. Sapiro, M. Jenkinson, D. A. Feinberg, E. Yacoub, C. Lenglet, D. C. V. Essen, K. Ugurbil, and T. E. Behrens, “Advances in diffusion mri acquisition and processing in the human connectome project”, NeuroImage, vol. 80, pp. 125–43, 2013.
[3] H. Y. Mesri, S. David, M. A. Viergever, and A. Leemans, “The adverse effect of gradient nonlinearities on diffusion mri: From voxels to group studies”, NeuroImage, vol. 205, pp. 116–27, 2019.
[4] J. Jovicich, S. Czanner, D. Greve, E. Haley, A. van der Kouwe, R. Gollub, D. Kennedy, F. Schmitt, G. Brown, J. MacFall, B. Fischl, and A. Dale, “Reliability in multi-site structural mri studies: Effects of gradient non-linearity correction on phantom and human data”, NeuroImage, vol. 30, no. 2, pp. 436–43, 2006.
[5] Z. Nagy, N. Weiskopf, D. C. Alexander, and R. Deichmann, “A method for improving the performance of gradient systems for diffusion-weighted mri”, Magnetic Resonance in Medicine, vol. 58, no. 4, pp. 763–8, 2007.
[6] S. Mohammadi, Z. Nagy, H. E. Möller, M. R. Symms, D. W. Carmichael, O. Josephs, and N. Weiskopf, “The effect of local perturbation fields on human dti: Characterisation, measurement and correction”, NeuroImage, vol. 60, no. 1, pp. 562–70, 2012.
[7] M. Paquette, C. Eichner, and A. Anwander, “Gradient non-linearity correction for spherical mean diffusion imaging”, Proc. Int. Soc. Magn. Reson. Med., vol. 27, no. 550, 2019.
[8] D. Topgaard, “Multidimensional diffusion mri”, Journal of Magnetic Resonance, vol. 275, pp. 98–113, 2017.
[9] C.-F. Westin, H. Knutsson, O. Pasternak, F. Szczepankiewicz, E. Özarslan, D. van Westen, C. Mattisson, M. Bogren, L. J. O’Donnell, M. Kubicki, D. Topgaard, and M. Nilsson, “Q-space trajectory imaging for multidimensional diffusion mri of the human brain”, NeuroImage, vol. 135, pp. 345–62, 2016.
[10] J. Sjölund, F. Szczepankiewicz, M. Nilsson, D. Topgaard, C.-F. Westin, and H. Knutsson, “Constrained optimization of gradient waveforms for generalized diffusion encoding”, Journal of Magnetic Resonance, vol. 261, pp. 157–6, 2015.
[11] F. Szczepankiewicz, C.-F. Westin, and M. Nilsson, “Maxwell-compensated design of asymmetric gradient waveforms for tensor-valued diffusion encoding”, Magnetic Resonance in Medicine, vol. 82, pp. 1424–37, 2019.
Figures