4408
General tools for diffusion tensor distributions, and matrix-variate Gamma approximation for multidimensional diffusion MRI data1Physical Chemistry, Lund University, Lund, Sweden, 2Random Walk Imaging AB, Lund, Sweden
Synopsis
Either on the voxel scale or within sub-voxel diffusion compartments, tissue microstructure can be described using a diffusion tensor distribution $$$\mathcal{P}(\mathbf{D})$$$. One way to resolve microstructural heterogeneity relies on choosing a plausible parametric functional form to approximate $$$\mathcal{P}(\mathbf{D})$$$. However, such a high-dimensional mathematical object is usually intractable. Here, we define matrix moments enabling the computation of diffusion metrics for any arbitrary functional choice approximating $$$\mathcal{P}(\mathbf{D})$$$. Applying these general tools to the matrix-variate Gamma distribution on the voxel scale, we obtain a new signal representation, the matrix-variate Gamma approximation, that we validate in vivo and in silico.
Introduction
The millimeter-scale resolution of diffusion MRI encompasses multiple microstructural tissue types, which implies that the diffusion signal probes a collection of microscopic diffusion profiles. Mathematically, this collection is described by a diffusion tensor distribution (DTD) $$$\mathcal{P}(\mathbf{D})$$$,1 yielding the diffusion signal$$\frac{\mathcal{S}(\mathbf{b})}{\mathcal{S}_0}=\int_{\mathrm{Sym}^+(3)}\!\mathcal{P}(\mathbf{D})\,\exp(-\mathbf{b}:\mathbf{D})\,\mathrm{d}\mathbf{D}=\left\langle\exp(-\mathbf{b}:\mathbf{D})\right\rangle\,,$$where $$$\mathbf{b}$$$ is the encoding tensor,2-4 $$$\mathrm{Sym}^+(3)$$$ denotes the space of 3x3 symmetric positive-definite matrices, ":" is the Frobenius inner product, and $$$\langle\cdot\rangle$$$ is the voxel-scale average. While nonparametric estimation of the DTD's features may be achievable,5-7 an alternative approach consists in approximating $$$\mathcal{P}(\mathbf{D})$$$ with a plausible parametric functional form whose parameters can be fitted against the acquired signals. This second approach, first implemented with distributions of scalar diffusivities,8-10 has recently been extended to DTDs in two ways:- the covariance tensor approximation,11 a two-term cumulant approach equivalent to considering a voxel-scale matrix-variate Gaussian (mv-Gaussian) DTD.12
- the DIAMOND model,13-15 a compartmental model wherein each anisotropic sub-voxel compartment is described by a matrix-variate Gamma (mv-Gamma) DTD.12
In this work, we first derive general tools facilitating the implementation of any functional choice for $$$\mathcal{P}(\mathbf{D})$$$. We then apply these tools to the mv-Gamma distribution and develop a new signal representation that describes the voxel content with a single mv-Gamma DTD: the "matrix-variate Gamma approximation". Finally, we validate this approximation in vivo and in silico.
Theory - Matrix moments of diffusion tensor distributions
We denote by "$$$\cdot$$$" and "$$$\otimes$$$" the matrix product and outer tensor product, respectively, with $$$\mathbf{D}^{\otimes\,\!2}\equiv\mathbf{D}\otimes\mathbf{D}$$$. Considering an arbitrary $$$\mathcal{P}(\mathbf{D})$$$, its moment-generating function writes12$$\mathcal{M}(\mathbf{Z})=\int\!\mathcal{P}(\mathbf{D})\,\exp\!\left[\mathrm{Tr}(\mathbf{Z}\cdot\mathbf{D})\right]\,\mathrm{d}\mathbf{D}=\left\langle\exp\!\left[\mathrm{Tr}(\mathbf{Z}\cdot\mathbf{D})\right]\right\rangle\,,$$for $$$\mathbf{Z}$$$ such that $$$\mathcal{M}(\mathbf{Z})$$$ is well-defined. $$$\mathcal{M}(\mathbf{Z})$$$ relates to the diffusion signal as15$$\mathcal{S}(\mathbf{b})=\mathcal{S}_0\,\mathcal{M}(-\mathbf{b})\,.$$By analogy with the moment-generating function of any scalar distribution, we showed that the first and second matrix derivatives of $$$\mathcal{M}(\mathbf{Z})$$$ yield the mean diffusion tensor $$$\langle\mathbf{D}\rangle$$$ and the fourth-order covariance tensor $$$\mathbb{C}=\langle\mathbf{D}^{\otimes\,\!2}\rangle-\langle\mathbf{D}\rangle^{\otimes\,\!2}$$$ associated to $$$\mathcal{P}(\mathbf{D})$$$. Indeed, we computed these derivatives using previous works16-18 as $$$\partial\mathcal{M}/\partial\mathbf{Z}=\left\langle\mathrm{exp}\!\left[\mathrm{Tr}(\mathbf{Z}\cdot\mathbf{D})\right]\,\mathbf{D}\right\rangle$$$ and $$$\partial^2\mathcal{M}/\partial\mathbf{Z}^2=\left\langle\mathrm{exp}\!\left[\mathrm{Tr}(\mathbf{Z}\cdot\mathbf{D})\right]\,\mathbf{D}\otimes\mathbf{D}\right\rangle$$$, deducing$$\langle\mathbf{D}\rangle=\left.\frac{\partial\mathcal{M}}{\partial\mathbf{Z}}\right|_{\mathbf{Z}=\mathbf{0}}\qquad\mathrm{and}\qquad\mathbb{C}=\left.\frac{\partial^2\mathcal{M}}{\partial\mathbf{Z}^2}\right|_{\mathbf{Z}=\mathbf{0}}-\left.\frac{\partial\mathcal{M}}{\partial\mathbf{Z}}\right|_{\mathbf{Z}=\mathbf{0}}^{\otimes\!\,2}\,.$$Additional efforts would yield the sixth-order skewness tensor11 $$$\mathbb{S}=\langle\mathbf{D}^{\otimes\,\!3}\rangle-3\langle\mathbf{D}\rangle\otimes\langle\mathbf{D}^{\otimes\!\,2}\rangle+2\langle\mathbf{D}\rangle^{\otimes\!\,3}$$$ via $$$\partial^3\mathcal{M}/\partial\mathbf{Z}^3$$$. While measures of voxel-scale diffusivity and anisotropy are extracted from $$$\langle\mathbf{D}\rangle$$$, measures of sub-voxel anisotropy and sub-voxel variance in isotropic diffusivities are obtained from $$$\mathbb{C}$$$ according to previous work.11Validation: We used the above formulae to retrieve the known mean tensor $$$\mathbf{M}$$$ and covariance tensor $$$\mathbf{\Sigma}\otimes\mathbf{\Psi}$$$ of the mv-Gaussian distribution12$$\mathcal{P}_\mathrm{Gauss}(\mathbf{X})=\frac{1}{(2\pi)^{9/2}\,\mathrm{Det}(\mathbf{\Sigma})^{3/2}\,\mathrm{Det}(\mathbf{\Psi})^{3/2}}\,\exp\left[\mathrm{Tr}\left(-\frac{1}{2}\,\mathbf{\Sigma}^{-1}\cdot(\mathbf{X}-\mathbf{M})\cdot\mathbf{\Psi}^{-1}\cdot(\mathbf{X}-\mathbf{M})^\mathrm{T}\right)\right]\,,$$for $$$\mathbf{X},\mathbf{M}$$$ arbitrary 3x3 real matrices, and $$$\mathbf{\Sigma},\mathbf{\Psi}\in\mathrm{Sym}^+(3)$$$, given its moment-generating function12$$\mathcal{M}_\mathrm{Gauss}(\mathbf{Z})=\exp\left[\mathrm{Tr}\left(\mathbf{Z}\cdot\mathbf{M}+\frac{1}{2}\,\mathbf{Z}^\mathrm{T}\cdot\mathbf{\Sigma}\cdot\mathbf{Z}\cdot\mathbf{\Psi}\right)\right]\,.$$
Theory - Matrix-variate Gamma approximation
We considered the non-central mv-Gamma distribution12 used in DIAMOND:14,15$$\mathcal{P}_\Gamma(\mathbf{D})=\frac{\mathrm{Det}(\mathbf{D})^{\kappa-2}}{\mathrm{Det}(\mathbf{\Psi})^\kappa\,\Gamma_3(\kappa)}\,\exp\!\left[-\mathrm{Tr}(\mathbf{\Theta}+\mathbf{\Psi}^{-1}\cdot\mathbf{D})\right]\,\mathcal{F}_{0,1}(\kappa,\mathbf{\Theta}\cdot\mathbf{\Psi}^{-1}\cdot\mathbf{D})\,,$$where $$$\mathbf{D}\in\mathrm{Sym}^+(3)$$$, $$$\kappa\in\;]1,+\infty[$$$, $$$\mathbf{\Psi}\in\mathrm{Sym}^+(3)$$$, and $$$\mathbf{\Theta}\in \mathrm{Sym}(3)$$$, with $$$\mathrm{Sym}(3)$$$ denoting the space of 3x3 symmetric matrices. $$$\Gamma_3$$$ is the multivariate Gamma function and $$$\mathcal{F}_{0,1}$$$ is the hypergeometric function of matrix argument of order $$$(0,1)$$$. The moment-generating function $$$\mathcal{M}_\Gamma(\mathbf{Z})$$$ of $$$\mathcal{P}_\Gamma$$$ is defined as12,15$$\mathcal{M}_\Gamma(\mathbf{Z})=\left[\mathrm{Det}(\mathbf{I}_3-\mathbf{Z}\cdot\mathbf{\Psi})\right]^{-\kappa}\exp\!\left[\mathrm{Tr}\!\left(\left[(\mathbf{I}_3-\mathbf{Z}\cdot\mathbf{\Psi})^{-1}-\mathbf{I}_3\right]\cdot\mathbf{\Theta}\right)\right]$$for $$$\mathbf{Z}\in\mathrm{Sym}(3)$$$ such that $$$(\mathbf{I}_3-\mathbf{Z}\cdot\mathbf{\Psi})\in\mathrm{Sym}^+(3)$$$, with the 3x3 identity matrix $$$\mathbf{I}_3$$$. The diffusion signal associated to $$$\mathcal{P}_\Gamma$$$ is given by15$$\mathcal{S}_\Gamma(\mathbf{b})=\mathcal{S}_0\,\mathcal{M}_\Gamma(-\mathbf{b})=\mathcal{S}_0\,[\mathrm{Det}(\mathbf{I}_3+\mathbf{\Psi}\cdot\mathbf{b})]^{-\kappa}\exp\!\left[-\mathbf{b}:[(\mathbf{I}_3+\mathbf{\Psi}\cdot\mathbf{b})^{-1}\cdot\mathbf{\Psi}\cdot\mathbf{\Theta}]\right]\,.$$Using our expressions for the matrix moments, we retrieved the mean diffusion tensor of $$$\mathcal{P}_\Gamma$$$ (employed in DIAMOND),14,15 and computed for the first time its covariance tensor, yielding$$\langle\mathbf{D}\rangle=\mathbf{\Psi}\cdot[\kappa\mathbf{I}_3+\mathbf{\Theta}]\qquad\mathrm{and}\qquad\mathbb{C}=\langle\mathbf{D}\rangle\otimes\mathbf{\Psi}+\mathbf{\Psi}\otimes\langle\mathbf{D}\rangle-\kappa\,\mathbf{\Psi}\otimes\mathbf{\Psi}\,.$$ The expression of $$$\langle\mathbf{D}\rangle$$$ and the symmetry of the different tensors imply that $$$\mathbf{\Psi}$$$ and $$$\mathbf{\Theta}$$$ share the same eigenvectors. Therefore, fitting $$$\mathcal{S}_\Gamma$$$ requires 11 parameters: $$$\mathcal{S}_0$$$, $$$\kappa$$$, the eigenvalues for $$$\mathbf{\Psi}$$$ and $$$\mathbf{\Theta}$$$, and the Euler angles orienting $$$\mathbf{\Psi}$$$'s eigenvectors. If numerically non positive-semidefinite, $$$\mathbb{C}$$$ is substituted with the nearest symmetric positive-semidefinite tensor.19Validation of the matrix-variate Gamma approximation
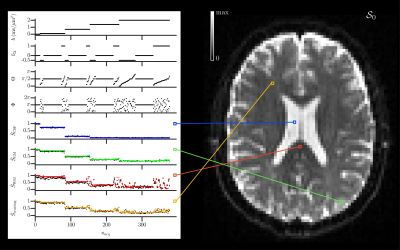
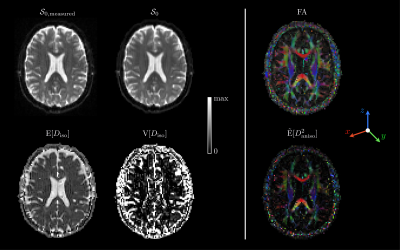
Statistical descriptors of interest: The mean diffusivity $$$\mathrm{E}[D_\mathrm{iso}]$$$, variance of isotropic diffusivities $$$\mathrm{V}[D_\mathrm{iso}]$$$, and normalized mean anisotropy $$$\tilde{\mathrm{E}}[D_\mathrm{aniso}^2]=\mathrm{E}[(D_\mathrm{iso}D_\Delta)^2]/\mathrm{E}[D_\mathrm{iso}]^2$$$ (similar to the microscopic $$$\mathrm{FA}$$$),10 with the isotropic diffusivity $$$D_\mathrm{iso}$$$, normalized diffusion anisotropy $$$D_\Delta\in[-0.5,1]$$$, and the voxel-scale expectation and variance $$$\mathrm{E}[\cdot]$$$ and $$$\mathrm{V}[\cdot]$$$, respectively.In vivo human brain: After implementing an iterative fitting of $$$\mathcal{S}_\Gamma$$$ in Matlab,20 we applied it to a dataset available online,21 which signal-to-noise ratio (SNR) was around 30 in the corona radiata.22 Fig.1 presents the acquisition scheme and shows that mv-Gamma reasonably fits the signals arising from diverse voxel contents. Fig.2 features consistent estimated parameter maps of $$$\mathcal{S}_0$$$, $$$\mathrm{E}[D_\mathrm{iso}]$$$, $$$\mathrm{V}[D_\mathrm{iso}]$$$ and $$$\mathrm{FA}$$$. However, $$$\tilde{\mathrm{E}}[D_\mathrm{aniso}^2]$$$ vanishes in areas of crossing fibers.
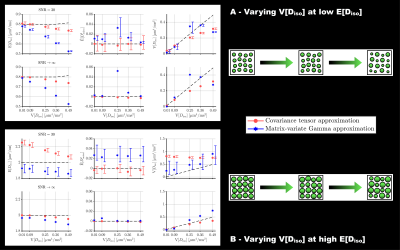
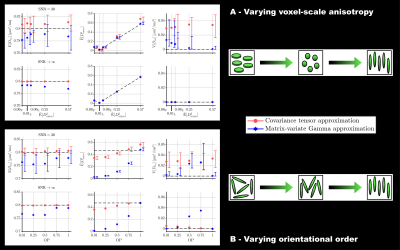
In silico: Various voxel contents were simulated in terms of distributions of $$$D_\mathrm{iso}$$$, $$$D_\Delta$$$ and orientations, with identical in vivo acquisition scheme and SNR. Medians and interquartile ranges on estimating $$$\mathrm{E}[D_\mathrm{iso}]$$$, $$$\tilde{\mathrm{E}}[D_\mathrm{aniso}^2]$$$ and $$$\mathrm{V}[D_\mathrm{iso}]$$$ were assessed across 100 Rician-noise realizations of the covariance tensor approximation (Cov)11 and the matrix-variate Gamma approximation (MV-Gamma), and compared to the simulated ground truth in isotropic (Fig.3) and anisotropic (Fig.4) systems. MV-Gamma adequately captures voxel-scale anisotropy and shows superior performance to Cov in accurately capturing $$$\mathrm{V}[D_\mathrm{iso}]$$$, but is inferior in estimating $$$\mathrm{E}[D_\mathrm{iso}]$$$. Besides, it cannot capture sub-voxel anisotropy ($$$\tilde{\mathrm{E}}[D_\mathrm{aniso}^2]$$$) in anisotropic systems with low orientational order, thereby confirming the previous in vivo observations.
Conclusion
We defined matrix moments enabling the computation of diffusion metrics for any functional choice approximating a diffusion tensor distribution $$$\mathcal{P}(\mathbf{D})$$$. Applying these tools to the mv-Gamma distribution up to its second matrix moment yields a new signal representation, the matrix-variate Gamma approximation, that performs adequately in orientationally ordered anisotropic systems (further justifying its use within DIAMOND),14,15 but misses sub-voxel anisotropy in systems with low orientational order. This missing information is likely contained in the sixth-order skewness tensor, which can be obtained via a third matrix moment.Acknowledgements
This work was financially supported by the Swedish Foundation for Strategic Research (AM13-0090, ITM17-0267) and the Swedish Research Council (2018-03697).References
1. Jian B et al., A novel tensor distribution model for the diffusion-weighted MR signal. Neuroimage 2007;37(1):164-176
2. Eriksson S et al., Isotropic diffusion weighting in PGSE NMR by magic-angle spinning of the q-vector. Journal of Magnetic Resonance 2013;226:13-18.
3. Eriksson S et al., NMR diffusion-encoding with axial symmetry and variable anisotropy: Distinguishing between prolate and oblate microscopic diffusion tensors with unknown orientation distribution. The Journal of Chemical Physics 2015;142(10):104201.
4. Topgaard D, Multidimensional diffusion MRI. Journal of Magnetic Resonance 2017; 275:98-113.
5. de Almeida Martins J P and Topgaard D, Two-dimensional correlation of isotropic and directional diffusion using NMR. Physics Review Letters 2016;116:087601.
6. de Almeida Martins J P and Topgaard, D Multidimensional correlation of nuclear relaxation rates and diffusion tensors for model-free investigations of heterogeneous anisotropic porous materials. Scientific Reports 2018;8,2488.
7. Topgaard D. Diffusion tensor distribution imaging. NMR in Biomedicine 2019;32(5):e4066
8. Yablonskiy D A et al., Statistical model for diffusion attenuated MR signal, Magnetic Resonance in Medicine 2003;50:664.
9. Jbabdi S et al., Model-based analysis of multishell diffusion MR data for tractography: How to get over fitting problems. Magnetic Resonance in Medicine 2012;68:1846.
10. Lasic S et al., Microanisotropy imaging: quantification of microscopic diffusion anisotropy and orientational order parameter by diffusion MRI with magic-angle spinning of the q-vector. Frontiers in Physics 2014;2:11.
11. Westin C-F et al. Q-space trajectory imaging for multidimensional diffusion MRI of the human brain. NeuroImage 2016;135:345-362.
12. Guptar A K and Nagar D K, Matrix Variate Distributions. Edited by Chapman and Hall/CRC, Boca Raton, Florida, 2000.
13. Scherrer B et al., Characterizing brain tissue by assessment of the distribution of anisotropic microstructural environments in diffusion-compartment imaging (DIAMOND). Magnetic Resonance in Medicine 2016;76:963.
14. Scherrer B et al., Decoupling axial and radial tissue heterogeneity in diffusion compartment imaging. In: Information Processing in Medical Imaging (IPMI) 2017;440-452.
15. Reymbaut A et al., The “Magic DIAMOND" method: probing brain microstructure by combining b-tensor encoding and advanced diffusion compartment imaging. In: International Society for Magnetic Resonance Imaging (ISMRM) 2018.
16. Brewer J, Kronecker products and matrix calculus in system theory. IEEE Transactions on Circuits and Systems 1978;25:772.
17. Laue S et al., Computing higher order derivatives of matrix and tensor expressions. In: Advances in Neural Information Processing Systems (NIPS) 2018.
18. Laue S et al., Matrix calculus, www.matrixcalculus.org. Accessed: 2019-07-30.
19. Higham N J, Computing a nearest symmetric positive semidefinite matrix. Linear algebra and its applications 1988;103:103-118.
20. Nilsson M et al., An open-source framework for analysis of multidimensional diffusion MRI data implemented in MATLAB. In: International Society for Magnetic Resonance Imaging (ISMRM) 2018.
21. Szczepankiewicz F et al., Linear, planar and spherical tensor-valued diffusion MRI data by free waveform encoding in healthy brain, water, oil and liquid crystals. Data in Brief 2019;25:104208.
22. Szczepankiewicz F et al., Tensor-valued diffusion encoding for diffusional variance decomposition (DIVIDE): Technical feasibility in clinical MRI systems. PLoS ONE 2019;14(3):e0214238.
Figures