4349
Automated Identification of Non-Brain Voxels for Clean Brain Extraction Using Diffusion MRI1Department of Radiology and BRIC, University of North Carolina, Chapel Hill, Chapel Hill, NC, United States
Synopsis
Automated brain extraction using diffusion MRI is challenging owing to the low spatial resolution. Unremoved residual non-brain voxels characteristically manifest as voxels with high fractional anisotropy (FA). In this abstract, we introduce a fast and robust method to identify non-brain voxels for clean extraction of the brain using diffusion MRI. We show that our method is effective for both adult and infant data.
Purpose
In diffusion MRI (dMRI), brain extraction is often performed by a simple thresholding of the non-diffusion-weighted image or with the help of the corresponding T1-weighted image. However, these approaches often result in residual unremoved non-brain voxels that exhibit characteristically high FA values. In this abstract, we introduce a method for clean extraction of the brain using diffusion MRI without relying on anatomical T1-weighted images.Methods
A spherical mean image is computed for each b-value by averaging across all images with the same b-value. We note that the spherical mean signal is not dependent on the fiber orientation distribution and is a function of per-axon diffusion characteristics1. Then $$$k$$$-means clustering is employed to partition the voxels into $$$k=2$$$ clusters representing the brain and non-brain voxels. Finally, median filtering and morphological closing are applied to remove isolated voxels and large holes, respectively. Then, the largest connected component is identified as the final mask.Materials
We evaluated our method using both adult and infant data. Adult: Multi-shell dMRI data from the Human Connectome Project (HCP)2, with $$$b=0, 1000, 2000, 3000\,\text{s}/\text{mm}^2$$$, voxel-size $$$1.25 \times 1.25 \times 1.25\,\text{mm}^3$$$.Infant: Multi-shell dMRI data of 16-days-old and 20-days-old infants from the Baby Connectome Project (BCP)3, $$$b=0, 500, 1000, 1500, 2000, 2500, 3000\,\text{s}/\text{mm}^2$$$, voxel-size $$$1.5 \times 1.5 \times 1.5\,\text{mm}^3$$$.
Results
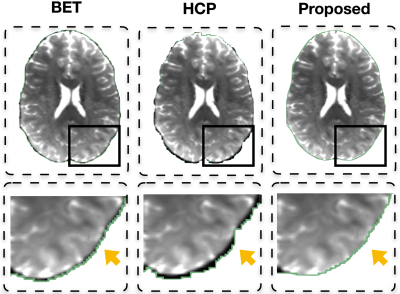
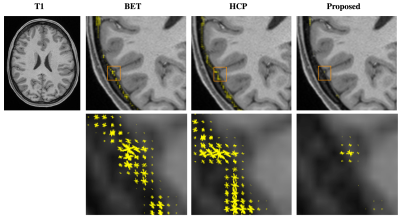
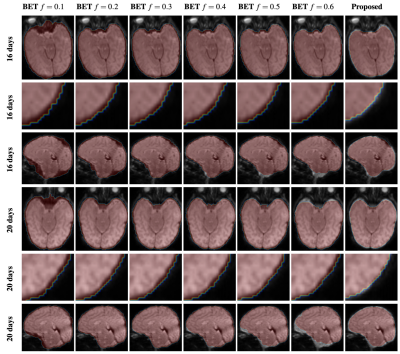
Figure 1 shows the adult brain extracted using BET4 , the HCP pipeline2 , and the proposed method. One can appreciate that with the proposed method cleaner brain extraction can be obtained (see yellow arrow). Table 1 shows the number of outlier voxels with FA values exceeding 1. Figure 2 shows the fiber orientation distributions of these outlier voxels overlaid on the T1-weighted image. These results further confirm the ability of the proposed method in cleanly extracting the brain. Moreover, our method requires only 2.33s on a single-core CPU and in hence significantly faster than BET (3.12s) and HCP brain extraction (31.89s). Figure 3 shows that our method identifies more effectively the expected brain voxels in the infant brains when compared with BET with increasing fractional intensity threshold.Acknowledgements
This work was supported in part by NIH grants (NS093842 and 1U01MH110274) and the efforts of the UNC/UMN Baby Connectome Project Consortium.References
1. Kaden E, Kelm ND, Carson RP, Does MD, Alexander DC. Multi-compartment microscopic diffusion imaging. NeuroImage. 2016;139:346-59.
2. Van Essen DC, Smith SM, Barch DM, Behrens TE, Yacoub E, Ugurbil K, Wu-Minn HCP Consortium. The WU-Minn human connectome project: an overview. Neuroimage. 2013;80:62-79.
3. Howell BR, Styner MA, Gao W, Yap PT, Wang L, Baluyot K, Yacoub E, Chen G, Potts T, Salzwedel A, Li G. The UNC/UMN baby connectome project (BCP): an overview of the study design and protocol development. NeuroImage. 2019;185:891-905.
4. Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. Fsl. Neuroimage. 2012;62(2):782-90.
Figures