4319
Performance comparison of three b-value sampling schemes in multiple diffusion models, including DTI, DKI, NODDI and MAP-MRI1MR Scientific Marketing, Siemens Healthcare, Shanghai, China, 2The First Affiliated Hospital of Zhengzhou University, Zhengzhou, China, 3Shanghai Key Laboratory of Magnetic Resonance, East China Normal Univeristy, Shanghai, China
Synopsis
This study aimed to evaluate the performance of three b-value sampling schemes in calculating multiple diffusion models, including the DTI, DKI, NODDI and the newly proposed mean apparent propagator (MAP)-MRI models. The three schemes includes the conventional diffusion spectrum imaging (DSI) acquisition scheme based on Cartesian grid sampling in q-space, multi-shell sampling with the same (MDDW) or different (FREE) gradient directions in each shell. Each scheme supports the estimation of all the four models. The results showed that generally three schemes generated very similar parameters, and could be all used in future studies.
Introduction
Multiple diffusion models are available to detect the brain microstructure, including conventional Diffusion Tensor Imaging (DTI) model, and more advanced models such as the Diffusion Kurtosis Imaging (DKI) [1], Neutire Orientation Dispersion and Density Imaging (NODDI) [2] and newly proposed Mean Apparent Propagator (MAP)-MRI [3]. In previous studies, different sampling schemes of b-value were used for these models, and normally the scheme was optimized for one specific model. In this study, we aim to evaluate the schemes that could be simultaneously used for multiple diffusion models, and three b-value sampling schemes were considered, including the conventional diffusion spectrum imaging (DSI) acquisition scheme based on Cartesian grid sampling in q-space, multi-shell sampling with the same (MDDW) or different (FREE) gradient directions in each shell.Methods
The brain diffusion data using three sampling schemes, namely DSI, FREE and MDDW, were acquired on a 3 T MR scanner (MAGNETOM Prisma, Siemens Healthcare, Erlangen, Germany) for five healthy subjects. The acquisition parameters of the three schemes with similar acquisition times were as follows: 1) DSI, 2 b=0 and 98 diffusion gradient directions with bmax = 3000 s/mm2, diffusion time δ/Δ = 15.9/35.0 ms, scan time = 6 min 37 s; 2) FREE, b = 0, 1000, 2000 and 3000 s/mm2 with 30 different gradient directions in each shell respectively, δ/Δ = 15.9/35.0 ms, scan time = 6 min 42 s; 3) MDDW, b = 0, 1000 and 2000 s/mm2 with 64 same gradient directions in each shell, δ/Δ = 13.9/33.0 ms, scan time = 7 min 57 s. The other parameters were the same for the three schemes: GRAPPA = 2, slice acceleration factor = 2, slice thickness = 2.2 mm, voxel size = 2.0 × 2.0 × 2.2 mm3, and slice number = 60. The quantitative parameters of DTI, DKI, MAP-MRI and NODDI models were calculated using software developed in-house with Python, called NeuDiLab, which is based on an open-resource tool DIPY (Diffusion Imaging In Python, http://nipy.org/dipy) and AMICO (https://github.com/daducci/AMICO) [4]. The DTI parameters included the fractional anisotropy (FA), the mean diffusivity (MD-DTI). The DKI parameters included the mean kurtosis (MK) and the mean diffusivity (MD-DKI). The MAP-MRI parameters included the return to the origin probability (RTOP), the return to the axis probability (RTAP), the return to the plane probability (RTPP), the mean square displacement (MSD) q-space inverse variance (QIV) and the non-Gaussianity (NG). The NODDI parameters included the intra-cellular volume fraction (ICVF), the isotropic volume fraction (ISOVF) and the orientation dispersion index (ODI).The structural similarity (SSIM) index was used to evaluate the structure similarity of parameter maps from every pair of sampling schemes [5]. In addition, two region of interest (ROI) with > 100 voxels were drawn in the gray matter (GM) and the white matter (WM) regions, respectively. The mean value for each diffusion parameter, derived from DSI, FREE and MDDW models, in ROI was compared using the direct comparison and scatter plots.
Results
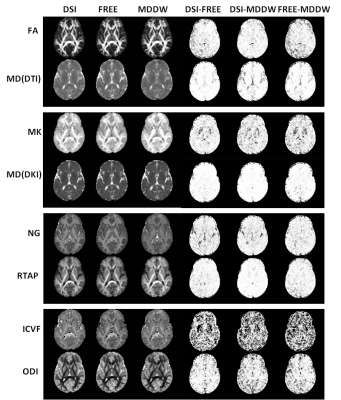
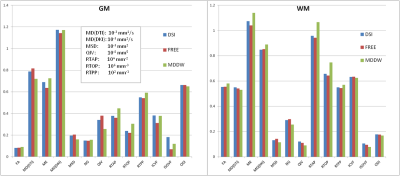
Figure 1 shows part parameter maps derived from the four diffusion models and their corresponding SSIM maps for the three sampling schemes. The parameter maps were very similar. Except NODDI, the SSIM maps from DTI, DKI and MAP-MRI models showed good agreements among the three schemes (Figure 1). Three schemes showed similar parameter values in both ROIs of GM and WM for the four models (Figure 2), and they also showed the good correlations for all the parameters, particularly for the DSI and FREE schemes (R2 = 0.988, p < 0.001) (Figure 3).Discussion
In the comparison of three sampling schemes using DTI, DKI, MAP-MRI and NODDI, the three schemes showed similar outcomes visually and quantitatively. Compared with the DSI and FREE schemes, the MDDW scheme showed slight larger differences in a few quantitative parameters of GM and WM, which might be due to the lower maximum b-value and lower number of gradient direction in MDDW scheme. Generally, three schemes generated very similar parameters, and could be all used in future studies. Meanwhile, considering potential application of the tractography, DSI and FREE schemes will be recommended as they used more gradient directions and could better solve the fiber crossing problem.Acknowledgements
No acknowledgement found.References
[1] Hui ES, et al. Neuroimage. 2015.
[2] Zhang H, et al. Neuroimage. 2012.
[3] Özarslan E, et al. NeuroImage. 2013.
[4] Daducci A, et al. NeuroImage. 2015.
[5] Wang Z, et al. IEEE Trans Image Processing. 2004.
Figures