4294
A Context-Aware Deep Attention Network for Thalamus Segmentation using 7T Multi-Modal MRI1Center for Magnetic Resonance Research, University of Minnesota, Minneapolis, MN, United States
Synopsis
In this study, we leverage 7T MR multi-modality and deep neural networks for accurate and efficient segmentation of the thalamus. Our contributions are 1) to build a dual-pathway and feature pyramid scheme to simultaneously encode global contextual information and local details within an encoder-decoder network; 2) to learn the optimal combination of global and local attentions to increase the feature representation power by adaptively recalibrating feature maps in an end-to-end manner. The proposed framework shows state-of-the-art performance on segmentation of the thalamus with 7T multi-modal MRI in an automatic and efficient way.
Introduction
The thalamus plays an essential role to relay sensory and motor signals between subcortical regions and the cerebral cortex, and is associated with neurodegenerative diseases and pathologies such as Alzheimer's disease, schizophrenia, and multiple sclerosis.1 Particularly, deep brain stimulation of the ventral intermediate nucleus within the thalamus has been shown to be effective for the treatment of essential tremor.2 Volumetric segmentation of the thalamus is thus a crucial step for diagnosis and treatment of such neurological disorders.3 Moreover, automatic segmentation facilitates clinical studies and neuro-modulation planning in terms of consistency and efficiency. Recent advances in 7 Tesla (T) MR imaging4–6 and computational potential of deep learning7–9 may allow automatic and fast processing in segmentation. However, MR imaging does not always clearly visualize the anatomically defined borders of the thalamus even though various modalities are available. Hence, developing an optimal combination of multiple contrasts to leverage complementary information on the images would improve segmentation. For volumetric segmentation in the medical domain, utilizing small sub-volumes (patches) for training within the network is typically considered to meet memory requirements and significantly increase the number of training samples.10 However, encoding features from such small patches may lead to missing sufficient contextual information for large structures during the learning phase. Further, an attention mechanism within the network makes features adaptively refined and thus boosts the feature representation power, which is useful for a reliable and interpretable system.11 In this study, to handle the above-mentioned issues, we propose a new attention-based deep learning architecture using 7T multi-modal MRI for accurate and efficient thalamus segmentation.Methods
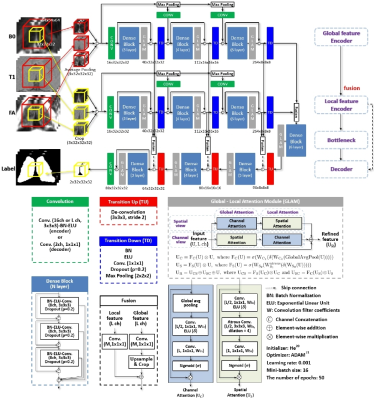
We incorporate the proposed context-aware and feature pyramid scheme (CAFP) into FC-DenseNet9 (Fig. 1). The first pathway involves learning high-level features such as the spatial and contextual information, while local details of target structures are learned in the second pathway. For a dense inference, feature maps from two pathways are spatially aligned and combined in fusion blocks and then integrated into the decoding path via the skip-connections. Furthermore, multi-scale input patches are concatenated into feature maps from transition down blocks for locality-aware learning. We also introduce the global-local attention module (GLAM) to highlight relevant features to segmentation (Fig. 1). To generate the optimal attention map, we propose to sequentially combine attention maps of a dual-pathway that are in channel attention first order and spatial attention first order, respectively, and finally add upon the given input feature maps. This module is incorporated into each convolutional dense block of the proposed network. We combine Tversky loss12 and focal loss13 for the training of the network. 7T multi-contrast MRIs (B0, T1-weighted, and fractional anisotropy (FA) images) of 43 subjects were jointly utilized in this study. Data acquisition protocols and pre-processing steps are detailed in the previous study.14 For each subject, B0 and FA images are co-registered to T1-weighted space for processing (resolution: 0.6×0.6×0.6mm3). The thalamus was manually segmented and served as ground truth for validation.15 To set the region of interest on a new input image, an atlas mask from a reference T1-weighted image from training data was linearly co-registered onto a T1-weighted test image. We compare the proposed network with a multi-atlas label fusion (STAPLE16) and commonly used deep neural networks - U-Net7, LiviaNet8, and FC-DenseNet9. Dice coefficient17 (DC), center of mass distance (CMD), mean surface distance (MSD) between ground truth and segmented results, and volumes are calculated for quantitative comparison.18 For statistical analysis of each measure, a one-way analysis of variance and Tukey’s honest significance post-hoc test were conducted for multiple comparisons. Five-fold cross-validation is used for evaluation.Results and Discussion
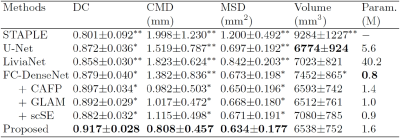
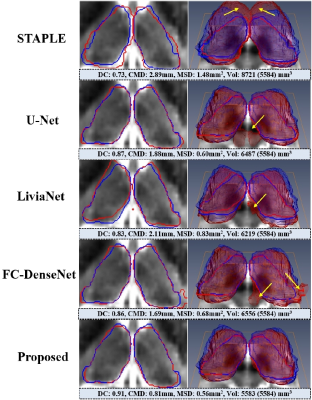
As shown in Fig. 2, the proposed network was the closest to the ground truth in terms of DC, CMD, and MSD. Deep neural network-based methods outperformed STAPLE by a large margin (p<0.001) with much faster inference (<30 sec on GPU). Such a large error and variance in STAPLE might be attributed to uncertainty in registration steps.18 The proposed network produced output with greater segmentation accuracy and consistency than U-Net and LiviaNet with fewer parameters, proving it is significantly more effective (p<0.001). Also, we can see the impact of each proposed component within the FC-DenseNet: the CAFP and the GLAM (improvement of 1.8% and 1.3%, respectively, in DC; p<0.05). Furthermore, we observed that the GLAM outperforms a state-of-the-art attention block (scSE19) within the FC-DenseNet consistently in each measure (p<0.05). Fig. 3 visualizes segmentation results on the 7T B0 MRI of a specific subject. Overall, the proposed network exhibited more comparable visualization to the ground truth than others, especially around the low contrast boundaries.Conclusion
In this study, we proposed a novel attention-based context-aware fully convolutional network for thalamus segmentation. A dual-pathway and feature pyramid scheme in the encoder was introduced for simultaneous learning of global and local features in an end-to-end manner. Also, we proposed to aggregate sequentially global and local attention maps both in channel and spatial viewpoints and integrate the attention module into the FC-DenseNet to increase the feature representation power. Experimental results demonstrate that the proposed network provides more accurate volumetric thalamus segmentation than current state-of-the-art approaches and can facilitate thalamus related studies in a fully automatic and efficient way.Acknowledgements
This work was supported in part by R01-NS081118, R01-NS113746, P50-NS098573, P30-NS076408 and P41-EB027061.References
1. Sherman, S. M. Thalamus. Scholarpedia 1, 1583
2. Papavassiliou, E. et al. Thalamic deep brain stimulation for essential tremor: relation of lead location to outcome. Neurosurgery 54, 1120–1130 (2004).
3. Coscia, D. M. et al. Volumetric and shape analysis of the thalamus in first-episode schizophrenia. Hum. Brain Mapp. 30, 1236–1245 (2009).
4. Abosch, A., Yacoub, E., Ugurbil, K. & Harel, N. An assessment of current brain targets for deep brain stimulation surgery with susceptibility-weighted imaging at 7 tesla. Neurosurgery 67, 1745–1756 (2010).
5. Cho, Z.-H. et al. Direct visualization of deep brain stimulation targets in Parkinson disease with the use of 7-tesla magnetic resonance imaging. J Neurosurg 113, 639–647 (2011).
6. Kerl, H. U. et al. The subthalamic nucleus at 7.0 Tesla: Evaluation of sequence and orientation for deep-brain stimulation. Acta Neurochir. (Wien). 154, 2051–2062 (2012).
7. Ronneberger, O., Fischer, P. & Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. in Proc. MICCAI 234–241 (2015).
8. Kamnitsas, K. et al. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal. 36, 61–78 (2017).
9. Jegou, S., Drozdzal, M., Vazquez, D., Romero, A. & Bengio, Y. The One Hundred Layers Tiramisu: Fully Convolutional DenseNets for Semantic Segmentation. in Proc. CVPR Workshop 11–19 (2017).
10. Bernal, J. et al. Quantitative Analysis of Patch-Based Fully Convolutional Neural Networks for Tissue Segmentation on Brain Magnetic Resonance Imaging. IEEE Access 7, 89986–90002 (2019).
11. Lee, H. et al. An explainable deep-learning algorithm for the detection of acute intracranial haemorrhage from small datasets. Nat. Biomed. Eng. (2018).
12. Sadegh, S., Salehi, M., Erdogmus, D. & Gholipour, A. Tversky loss function for image segmentation using 3D fully convolutional deep networks. in Proc. MICCAI Workshop (MLMI) 379–387 (2017).
13. He, K., Goyal, P., Girshick, R., Dollar, P. & Lin, T.-Y. Focal loss for dense object detection. IEEE Trans. Pattern Anal. Mach. Intell. to be Publ. (2018).
14. Plantinga, B. R. et al. Individualized parcellation of the subthalamic nucleus in patients with Parkinson’s disease with 7T MRI. Neuroimage 168, 403–411 (2016).
15. Duchin, Y. et al. Patient-specific Anatomical Model for Deep Brain Stimulation based on 7 Tesla MRI. PLoS One 13, 1–23 (2018).
16. Warfield, S. K., Zou, K. H. & Wells, W. M. Simultaneous truth and performance level estimation (STAPLE): An algorithm for the validation of image segmentation. IEEE Trans. Med. Imaging 23, 903–921 (2004).
17. Dice, L. R. Measures of the amount of ecologic association between species. Ecology 26, 297–302 (1945).
18. Kim, J. et al. Automatic localization of the subthalamic nucleus on patient-specific clinical MRI by incorporating 7 T MRI and machine learning: Application in deep brain stimulation. Hum. Brain Mapp. 40, 679–698 (2019).
19. Roy, A. G., Navab, N. & Wachinger, C. Recalibrating Fully Convolutional Networks With Spatial and Channel ‘Squeeze and Excitation’ Blocks. IEEE Trans. Med. Imaging 38, 540–549 (2019).
20. He, K., Zhang, X., Ren, S. & Sun, J. Delving Deep into Rectifiers : Surpassing Human-Level Performance on ImageNet Classification. in Proc. ICCV 1026–1034 (2015).
21. Kingma, D. P. & Ba, J. L. ADAM: a method for stochastic optimization. in Proc. ICLR (2015).
Figures