4160
Declutter the protocol tree: managing and comparing sequence parameters of multiple clinical systems using Python tools1Radiology, University Hospital Gent, Gent, Belgium, 2Ghent Institute for functional and Metabolic Imaging, Ghent University, Gent, Belgium
Synopsis
Monitoring protocol and sequence changes is of key importance for efficiently running a clinical imaging department. Since an MRI protocol contains hundreds of parameters, it is almost impossible to manage protocols from the scanner console, especially in a clinical situation where console time for the MR physicist is very limited. We present a python-based platform that performs all these duties and show 4 potential use cases: 1) compare parameters from sequences that share the same name 2) same purpose, different name 3) differences in naming of Regions/Exams/Programs 4) real-life example of decluttering a pediatric neuro protocol.
Introduction
Improving workflow and standardizing care are important factors in a clinical environment. Ideally, clinical MR protocols should be standardized over multiple scanners to provide the same diagnostic quality images. In real-life, a MR protocol tree is a dynamic thing; protocols are added, modified or discarded regularly, which may result in inconsistent protocols and decreasing quality. Monitoring these changes is of key importance for efficiently running a clinical imaging department. Since an MRI protocol contains hundreds of parameters, it is almost impossible to manage protocols from the scanner console, especially in a clinical situation where console time for the MR physicist is very limited. Although a number of vendors provide commercial solutions[1,2], these do not show all the parameters in a simple text view or lack the option of basic version control and diff operations. It is often needed to revert to an older version of a protocol, to see if a protocol has changed, or to check protocols from two different scanners. We present a Python-based platform that performs all these duties and show 4 potential use cases.Methods
On Siemens systems, protocols are organized in a protocol tree, then in Regions, Exams, and Programs. A Program contains all the sequences. Any of these can be printed to a xml file. We have developed Python scripts that parse these xml files, using the Python function "etree" from "lxml", and creates a dictionary of sequences including a unique sequence ID and a full set of parameters. It can compare sequences within one scanner, compare xml files from two scanners or find the differences between two xml files, i.e. before and after modification of a scan protocol, using Python's "xmldiff". One line per sequence that contains all parameters is written to a Python Pandas dataframe and csv text file for further analysis. It also creates easy to read Excel files which can be used by the MR physicist or radiographer to modify parameters at the scanner console. The initial xml print from the scanner is monitored for changes with git.Results
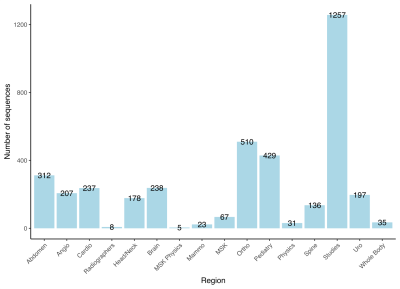
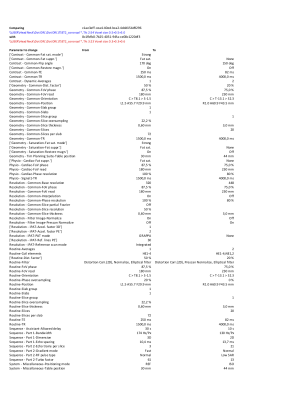
Figure 1 shows the scale of the problem. In this example from one clinical system, the xml file contained 120.4Mb of text data. There are 3870 sequences divided over 16 Regions. Per sequence, a maximum of 900 parameters are available.Use case 1 In this example we have compared the parameters of the sequences that have the same name. 65 duplicate sequences were found. In one example sequence, a coronal T2* TSE from the Head/Neck exam, there was a mismatch of 72 parameters, see figure 2.
Use case 2 In this example we looked for sequences that have the same purpose, but differ in name. The CSV output of the Python script was loaded into Tad v0.9.0 (https://www.tadviewer.com/) for easy display and filtering of the data. We can identify that there are multiple sequences for DWI or DTI of the brain (see figure 3), but that the resolution for these sequences is different. This may be on purpose, but has to be checked with radiologists and fixed on the scanner console (see case 4).
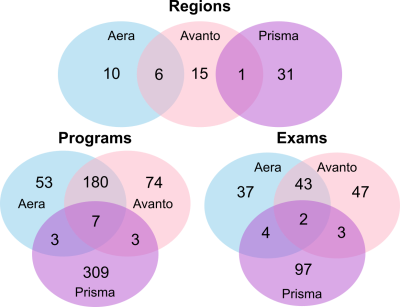
Use case 3 In this case we have explored what the differences in Region, Exam, and Program names are at three different scanners. The Venn diagram in figure 4 shows that there is a considerable difference in both the total number of Regions, Exams, and Programs, and that there is very small overlap between all three systems. This means that there is no uniform naming convention between the three systems, which should be addressed.
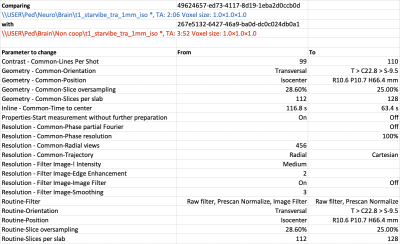
Use case 4 Here we provide a real-life example for a pediatric neuro exam. All sequences within a Program should have a unique name, such that the combination of Region, Program and sequence is unique. As a first step we have standardized sequence naming, following the convention <contrast>_<sequence type>_<orientation>_<dimension>_<optional: intended use>. Using this convention we can establish unique sequence names, i.e. t2fs_qtse_tra_3mm_0-2y which stands for a quiet axial t2 fatsat with a slice thickness of 3mm, for use in pediatric patients < 2y of age. After this step, MR_compare_sequences.py was executed. Out of 84 sequences in the neuro Program, 35 were duplicates and 7 duplicate sequences contained different parameters that needed to be adjusted on the scanner console (see figure 4). This whole process can be done by a MR technologist or physicist in a few hours.
Discussion and conclusion
Using the developed tools, an efficient workflow is created to manage protocol trees, which saves valuable console time at the scanner. We have demonstrated that the tools can pinpoint differences in sequence parameters and present them in a readable format. These tools can also be used for standardization in multi-center studies to check if sequence parameters have changed after sequence import.Release versions of the Python scripts will be made available through www.github.com/GIfMI
Acknowledgements
No acknowledgement found.References
1. Siemens Teamplay Protocols https://www.siemens-healthineers.com/digital-health-solutions/digital-solutions-overview/service-line-managment-solutions/teamplay/teamplay-performance-management-applications/teamplay-protocols, visited Oct 31, 2019
2. GE MR Excellence, GE Signa Pulse, vol. 26, Spring 2019
Figures