3992
Simulation of postsurgical functional connectome from presurgical resting-state fMRI: optimization and validation1Imaging Physics, UT MD Anderson Cancer Center, Houston, TX, United States, 2Neuroradiology, UT MD Anderson Cancer Center, Houston, TX, United States, 3Neurosurgery, UT MD Anderson Cancer Center, Houston, TX, United States, 4Neuropsychology, UT MD Anderson Cancer Center, Houston, TX, United States
Synopsis
Post glioma-resection functional connectomic measures were simulated by modifying the presurgical resting-state fMRI data using the resection margin in 13 patients. The simulation uses atlas based approaches that removes the signal contribution in the resected area based on the portion of parcel removed. The simulated connectomic measures were found to approximate the actual postsurgical connectomic measures. BN246 atlas appears to be more stable in predicting the changes in connectomic measures than AAL90 atlas. The results show that derivation of postsurgical functional connectomic measures from presurgical data is feasible.
Introduction
Gliomas represents approximately 80% of malignant brain tumors1. Treatment of glioma typically involve neurosurgical intervention (e.g. resection). Unfortunately the majority of patients experience postsurgical decline in neurocognitive function (NCF). Brain lesions directly impact the cerebral network organization and is associated with alterations in NCF. Studies in various neurological diseases have linked changes in the brain’s functional connectomic measures to NCF decline2-7. Simulation of disruption to brain network and the prediction of NCF may be especially useful in surgical planning. However, methods of simulation has not been well established and few studies have validated the simulation with post-lesioned brain8,9. We hypothesize that, given the limited neuroplasticity shortly after surgery (<4 weeks), functional connectivity measures in this period may be used to optimize and validate the simulated lesioning on the presurgical functional connectome using the resection margin.Methods
Resting-state (rs)-fMRI, 3D GRE T1-weighted and 2D fluid attenuated inversion recovery scans were acquired from 13 gliomas patients within 1 week before and 4 weeks after the tumor resection. Resected areas were outlined on presurgical T1w images by an experienced neuroradiologist by comparing the pre- and postsurgical structural MR images. These ROIs were then spatially transformed to the presurgical rs-fMRI images via registration between the T1w and EPI images. Pre- and postsurgical rs-fMRI data were pre-processed using a standard pipeline consisting of timing and motion correction, spatial normalization, regressing-out of covariates, and bandpass filtering implemented using MATLAB (MathWorks, Natick, MA) and SPM12 (Wellcome Department of Cognitive Neurology, Institute of Neurology, London, UK). AAL9010 and BN24611 atlas were used to extract the ROI time course to generate the pre- and postsurgical connectivity matrices using MATLAB. To simulate the postsurgical functional connectivity from the presurgical rs-fMRI data, signal in the resected were removed from the rs-fMRI data using two methods: 1) the signal from the resected area was set to zero, and 2) same as 1), but if the extent of the resection exceeds a percentage of the parcel, then the signal from the entire parcel was set to zero as well. For method 2), a threshold of 0%, 25%, 50%, and 75% were investigated. Several network measures were calculated over a range of connectivity density from 0.1 to 0.3 using the Brain Connectivity Toolbox12. To assess the performance of the simulations, paired t-test were used to identify if the difference between the simulated postsurgical measures and the actual postsurgical measures were significantly smaller than the difference between the actual pre- and postsurgical measures.Results
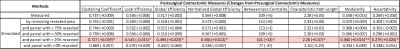
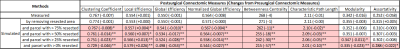
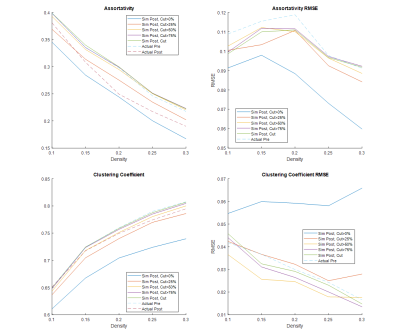
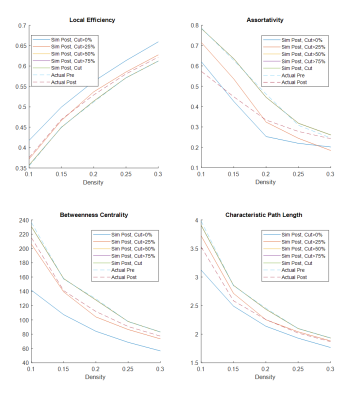
Table 1 and 2 show the postsurgical connectomic measures, their changes from pre- to postsurgery and whether the changes obtained by the simulations were significant for the two different atlases used. Using the AAL90 atlas, the best performing simulating was obtain using a parcel removal threshold of 25%, with significant results across all the calculated measures. Using the BN246 atlas, the best performing simulation was obtained using a parcel threshold of 0%, but other thresholds (25-75%) also had good performance in most connectomic measures. Figure 1 shows selected patient-average connectomic measures that illustrates the performances of different methods in simulating postsurgical connectomic measures.Discussion
The results here show that derivation of postsurgical functional connectivity from presurgical data is possible. Using a relatively simple atlas based approach we were able to approximate several postsurgical connectomic measures using only the presurgical data and resection margin as inputs, as illustrated in Figure 2 for one of the patients. Simulated connectomic measures obtained by simply removing the signal from the resected area resulted in postsurgical changes that is significantly different from the actual postsurgical data, likely because the remaining tissue is not a good functional representation of the parcels. By removing the signal from an entire parcels based on the percentage of the parcel resected, the simulation was significantly improved. It was interesting that the best performing parcel threshold was 25% using the AAL90 atlas and 0% using the BN246 atlas. This may be due to the difference in parcel size. BN246 atlas has nearly 3x smaller parcel on average than AAL90 atlas, and may therefore be more resilient to the node removal. The measures derived using BN246 atlas also appear to be more stable in predicting the changes in connectomic measures across the different thresholds. Testing the simulated connectomic measures with their association with the actual NCF changes is underway. Nevertheless, the results here points to the feasibility of simulating postsurgical functional connectivity using only the presurgical rs-fMRI data.Conclusion
The results here show that derivation of postsurgical functional connectomic measures from presurgical data is feasible.Acknowledgements
No acknowledgement found.References
1. Ostrom QT, Gittleman H, Truitt G, Boscia A, Kruchko C, Barnholtz-Sloan JS. CBTRUS Statistical Report: Primary brain and other central nervous system tumors diagnosed in the United States in 2011–2015. Neuro-oncology 2018 Oct 1;20(suppl_4):iv1-iv86.
2. Caeyenberghs K, Leemans A, De Decker C, Heitger M, Drijkoningen D, Vander Linden C, Sunaert S, Swinnen SP. Brain Connectivity and Postural Control in Young Traumatic Brain Injury Patients: A Diffusion MRI Based Network Analysis. NeuroImage: Clinical 2012; 1 (1): 106–15.
3. Caeyenberghs K, Leemans A, Leunissen I, Gooijers J, Michiels K, Sunaert S, Swinnen SP. Altered Structural Networks and Executive Deficits in Traumatic Brain Injury Patients. Brain Structure and Function 2014: 219 (1): 193–209.
4. Hellyer PJ, Scott G, Shanahan M, Sharp DJ, Leech R. Cognitive Flexibility through Metastable Neural Dynamics Is Disrupted by Damage to the Structural Connectome. Journal of Neuroscience 2015; 35 (24): 9050–63.
5. Fagerholm ED, Hellyer PJ, Scott G, Leech R, Sharp DJ. Disconnection of network hubs and cognitive impairment after traumatic brain injury. Brain 2015;138 (6): 1696–1709.
6. Yuan W, Wade SL, Babcock L. Structural connectivity abnormality in children with acute mild traumatic brain injury using graph theoretical analysis. Human Brain Mapping 2015; 36 (2): 779–92.
7. Huang Q, Zhang R, Hu X, Ding S, Qian J, Lei T, Cao X, Tao L, Qian Z, Liu H. Disturbed small-world networks and neurocognitive function in frontal lobe low-grade glioma patients. PLoS One 2014;9(4):e94095
8. Cabral J, Hugues E, Kringelbach ML, Deco G: Modeling the outcome of structural disconnection on resting-state functional connectivity. NeuroImage 2012; 62(3):1342–1353.
9. Hart MG, Price SJ, Suckling J: Connectome analysis for pre-operative brain mapping in neurosurgery. Br J Neurosurg 2016; 30(5):506–517.
10. Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 2002;15 (1): 273-89.
11. Fan L, Li H, Zhuo J, Zhang Y, Wang J, Chen L, Yang Z, Chu C, Xie S, Laird AR, Fox PT. The human brainnetome atlas: a new brain atlas based on connectional architecture. Cerebral cortex 2016; 26 (8): 3508-26.
12. Rubinov M, Sporns O. Complex Network Measures of Brain Connectivity: Uses and Interpretations. NeuroImage 2010; 52 (3): 1059–69.
Figures