3954
OpenMind – An Open Database of Brain Functions under Anesthesia for Preclinical MRI Research (and Beyond)1Berlin Ultrahigh Field Facility (B.U.F.F.), Max Delbrück Center for Molecular Medicine in the Helmholtz Association, Berlin, Germany, 2Unaffiliated, Montreal, QC, Canada, 3Experimental and Clinical Research Center, a joint cooperation between the Charité Medical Faculty and the Max Delbrück Center for Molecular Medicine in the Helmholtz Association, Berlin, Germany
Synopsis
Understanding anesthetic effects in preclinical fMRI is key to produce significant findings. Over the last decades massive amounts of data have been produced on anesthesia, yet major portions lie idle to the community. Here we introduce an open, community driven infrastructure that combines the wiki principle (dynamic creating, correcting, commenting) with a powerful dynamic databank approach (fast and easy search, and network visualization of related findings) on anesthetic effects. OpenMind provides (1) convenient data upload, (2) fast and efficient search and filter
functions, (3) embedding of findings in a larger dynamic context, and
(4) low maintenance.
Introduction
In preclinical functional MRI anesthesia is commonly applied. Growing evidence reveals the shortcomings of classic monoanesthetic approaches1-5. The aim of sufficiently sedating the animal yet preserving whole-brain functionality is the enigmatic bottleneck for generating meaningful findings of translational impact1. A comprehensive understanding of anesthetic mechanisms is key to produce reliable and significant research. Over the last decades massive amounts of data on effects of various anesthetics have been produced across species and modalities ranging from single neuron recordings to whole-brain functional connectivity1-8. Yet major portions of this knowledge are fragmented over disparate domains or lie idle to the community – partially due to the sheer amount of data, partially due to a no-reward policy for communicating “negative” or “low-impact” results, which causes severe loss of valuable information on anesthetic (side) effects and reproducibility which provides a major obstacle in translational research. Gathering, mining and classifying these data to make them accessible and easy to find would be of highest relevance, value and utility. Recognizing the challenges of such an endeavor, we propose an infrastructure to minimize the effort and maximize the gain of contributing in building a powerful open database of anesthetic effects on brain functions that provides (1) convenient data upload, (2) fast and efficient search and filter functions, (3) embedding of findings in a larger dynamic context, and (4) low maintenance.Methods
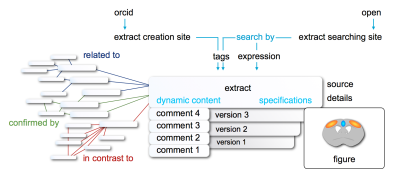
OpenMind uses Elasticsearch (elastic.co) to store and search all information entered by researchers. Elasticsearch provides a robust, fast, and flexible platform which is widely used and has been heavily tested by the data science community.The primary design goals of OpenMind are ease of use, reproducibility (of searches, links, etc.), and rigour. By focusing on small results, the barrier to publication is all but removed. An extract (defined below) can be created in a matter of minutes, comments, reviews, and other metadata can be added to extracts with similar ease, which in turn maintains rigour: an organic peer review process that augments the established, journal centered, peer review system. Changes to data are treated as additions, not replacements, and all previous versions of information are kept. Thus searches are genuinely reproducible, and references within the platform will never be invalidated by changes to the referent.
Results
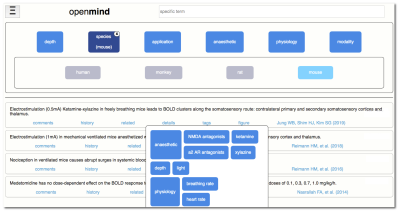
OpenMind deals in extracts: brief takeaways of about one sentence summarizing a single main finding – sources may include published articles, or unpublished lab notes, with one article potentially engendering many extracts (Fig. 1).A specified filter system permits to tag and find extracts based on their content, including anesthetic class and depth, species, modalities, applications, and physiology (Fig. 2). One can query, for instance, only extracts that report on effects of low doses of ketamine and xylazine in mice using fMRI with somatosensory stimuli. Search by expression across the library is also possible.
An essential feature of OpenMind is to generate dynamic context by connecting extracts, either ‘related to‘ (neutral), ‘confirmed by‘ or ‘in contrast to‘ other extracts. These relations make it easy to organize knowledge in a way that meets the requirements for contemporary research: capturing complex relations and contradicting findings in a fast and comprehensive way. A visualization option to display extract relation networks will be implemented, which allows for a comprehensive overview on a specific finding embedded in a dynamic scientific discourse that clarifies the big picture.
A major focus of the project is to keep these processes as swift and convenient as possible to make contributing a pleasant experience. Implemented features include a simple and intuitive interface, automated article recognition via pubmed link, and figure upload by drag-and-drop. Other features will follow along these lines.
Discussion
OpenMind is an open hub of scientific findings on anesthetic effects across depth and classes, species, monitoring techniques and modalities. It combines the wiki principle (dynamic creating, correcting, commenting by members of the community) with a powerful and dynamic databank approach (fast and easy search, network visualization of related findings). Scientific findings are isolated into key messages (so-called „extracts“) that are organized in a novel, intuitive, and easily accessible way. The opportunity to create extracts based on lab reports makes it from the very start a valuable source to share, find and compare unpublished data on anesthetic (side) effects. Particular care was taken to make the extract creation process a swift and pleasant experience – to lower the hurdle for every scientist to actively contribute, growing and expanding the network toward a reliable encyclopedia for anesthesia across fields.Conclusion
Motivated by the leading idea to share information by and with the community for mutual benefit, OpenMind is a three stage endeavor: (1) identifying the challenge and building an infrastructure, (2) contribution of pioneers who support the project by generating a critical mass of content and providing constructive feedback, and (3) going public and providing a powerful platform to benefit the community. After completion of the OpenMind development phase we are looking forward upon the faith and constructive feedback of the community. Most important for the MRI community, OpenMind provides a technological base for advancing functional MRI in basic neuroscience and translational research.Acknowledgements
OpenMind was born as an intellectual child of the MRathon, an hackathon for open science worldwide that was hosted in 2019 in Montreal as a satellite event to the ISMRM. We thank the organizers for bringing the team behind OpenMind together and the participants for their support and invaluable feedback.
References
1. Reimann & Niendorf (submitted). The (un)conscious mouse as a model for human brain functions: key principles of anesthesia and their impact on translational neuroimaging. Front Syst Neurosci.
2. Sharp, P.S., Shaw, K., Boorman, L., Harris, S., Kennerley, A.J., Azzouz, M., and Berwick, J. (2015). Comparison of stimulus-evoked cerebral hemodynamics in the awake mouse and under a novel anesthetic regime. Sci Rep 5, 12621.
3. Shim, H.-J., Jung, W.B., Schlegel, F., Lee, J., Kim, S., Lee, J., and Kim, S.-G. (2018). Mouse fMRI under ketamine and xylazine anesthesia: Robust contralateral somatosensory cortex activation in response to forepaw stimulation. NeuroImage 177, 30-44.
4. Lu, H., Zou, Q., Gu, H., Raichle, M.E., Stein, E.A., and Yang, Y. (2012). Rat brains also have a default mode network. Proc Natl Acad Sci U S A 109, 3979-3984.
5. Grandjean, J., Schroeter, A., Batata, I., and Rudin, M. (2014). Optimization of anesthesia protocol for resting-state fMRI in mice based on differential effects of anesthetics on functional connectivity patterns. Neuroimage 102 Pt 2, 838-847.
6. Paasonen, J., Stenroos, P., Salo, R.A., Kiviniemi, V., and Grohn, O. (2018). Functional connectivity under six anesthesia protocols and the awake condition in rat brain. Neuroimage 172, 9-20.
7. Grandjean, J. et al. (2019). Common functional networks in the mouse brain revealed by multi-centre resting-state fMRI analysis. bioRxiv, 541060.
8. Barttfeld, P., Uhrig, L., Sitt, J.D., Sigman, M., Jarraya, B., and Dehaene, S. (2015). Signature of consciousness in the dynamics of resting-state brain activity. Proc Natl Acad Sci U S A 112, 887-892.
Figures
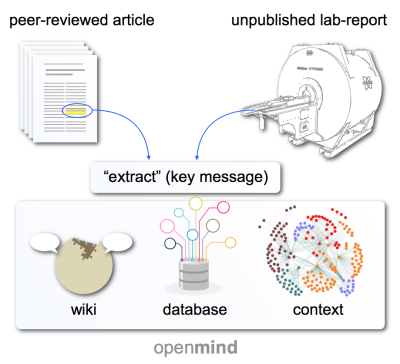
Fig. 1 OpenMind provides an infrastructure for sharing, searching, and contextualizing scientific findings around anesthetic effects. Findings are isolated into key messages (so-called extracts) that are extracted from peer-reviewed articles or unpublished lab-reports. The infrastructure combines elements from the wiki approach (creation, correction, discussion of dynamic content), dynamic database (fast and powerful search and ranking), and dynamic network analysis (creation of context and visualization of related findings) – all swift and easy to handle.