3932
Estimating Cerebral Blood Flow from BOLD Signal Using Deep Dilated Wide Activation Networks
Danfeng Xie1, Danfeng Xie1, Yiran Li1, Hanlu Yang1, Li Bai1, Donghui Song 2, Yuanqi Shang2, Qiu Ge2, and Ze Wang3
1Temple University, Philadelphia, PA, United States, 2Hangzhou Normal University, Hangzhou, China, 3University of Maryland School of Medicine, Philadelphia, MD, United States
1Temple University, Philadelphia, PA, United States, 2Hangzhou Normal University, Hangzhou, China, 3University of Maryland School of Medicine, Philadelphia, MD, United States
Synopsis
The purpose of this study was to synthesize Arterial Spin Labeling (ASL) cerebral blood flow (CBF) signal from blood-oxygen-level-dependent (BOLD) fMRI signal using deep machine learning (DL). Experimental results in the dual-echo Arterial Spin Labeling sequence show that the BOLD-to-ASL synthesize networks, the BOA-Net will yield similar cerebral blood flow value to that measured by ASL MRI and the cerebral blood flow maps produced by BOA-Net will show higher Signal-to-Noise Ratio (SNR) than that from ASL MRI.
Introduction
Arterial Spin Labeling (ASL) perfusion MRI [8] and blood-oxygen-level-dependent (BOLD) fMRI [7] provide complementary information for assessing brain functions. BOLD fMRI has higher signal-to-noise-ratio (SNR) and higher spatial temporal resolution than ASL but it only provides a relative measure. ASL is quantitative and it’s more clinically relevant for assessing neurologic or psychiatric diseases but has not been included in many fMRI experiments. The purpose of this study was to synthesize ASL cerebral blood flow (CBF) signal from BOLD signal using deep machine learning (DL). We hypothesize that the BOLD-to-ASL synthesize networks, the BOA-Net will yield similar CBF value to that measured by ASL MRI and the CBF maps produced by BOA-Net will show higher SNR than that from ASL MRI.Methods
Figure 1 shows the network architecture of BOA-Net. Two pathways were used. The global pathway was composed of the Dilated Convolution [10] where the receptive field of each neuron was dilated to learn more global data representations while the local pathway was to keep the local details. Both pathways used wide activation residual blocks to expand data features and pass more information through the network [3]. We then dubbed this dilated filters and wide activation blocks-based BOA-Net as the DWAN. ASL and BOLD fMRI data were acquired with the dual-echo ASL sequence [4] from 50 young healthy subjects. Imaging parameters were: labeling time/delay time/TR/TE1/ TE2 = 1.4s/1.5s/4.5s/11.7 msec/68msec, 90 acquisitions (90 BOLD images and 45 Control/Label (C/L) image pairs), FOV=22 cm, matrix=64x64, 16 slices with a thickness of 5 mm plus 1 mm gap. We used ASLtbx [6] to preprocess ASL images using the steps described in [5]. BOA-Net was trained with data from 23 subjects’ CBF maps (BOLD fMRI as input and ASL CBF as the training reference). 4 different subjects were used for validation samples. The remaining 23 subjects' CBF maps were used as test samples. To reduce memory demand, 2D BOA-Net was used in this study. The model was trained using the 7th to the 11th axial image slices, each with 64 x 64 pixels. BOA-Net was also implemented using U-Net [9] and DilatedNet [2], two popular CNN structures widely used in medical imaging, as comparisons to the DWAN-based BOA-Net. We use Peak signal-to-noise ratio (PSNR) and structure similarity index (SSIM) to quantitatively compare the performance of DWAN with U-Net and DilatedNet. SNR was used to measure image quality. It was calculated by using the mean signal of a grey matter region-of-interest (ROI) divided by the standard deviation of a white matter ROI in slice 9.Results
Our testing results demonstrated that ASL CBF can be reliably predicted from BOLD fMRI with comparable image quality and higher SNR. Figure 2 shows the data from a representative subject. All BOA-Nets (based on U-net, DilatedNet, and DWAN) yielded CBF maps with similar visual appearance. As compared to the genuine mean CBF map measured by the dual-echo ASL MRI sequence (surrogate groundtruth), the BOLD-derived CBF map produced by BOA-Net showed substantially improved quality in terms of suppressed noise and better perfusion contrast between grey matter and white matter. BOA-Net further recovered CBF signals in the air-brain boundaries where BOLD fMRI often suffers from signal loss due to the large susceptibility gradients. Figure 3 shows the notched box plot of SNR between surrogate groundtruth and the output of BOA-Net. the average SNR of surrogate groundtruth, DilatedNet-based BOA-Net, U-Net-based BOA-Net and DWAN-based BOA-Net was 7.56, 7.99, 13.12 and 13.33. BOA-Net achieved a 76.32% SNR improvement compared with the mean CBF maps of ASL. The PSNR of U-Net, DilatedNet and DWAN are 21.88, 21.51 and 22.49, respectively. the SSIM of U-Net, DilatedNet and DWAN are 0.856, 0.841 and 0.865, respectively. DWAN achieved higher PSNR and SSIM than U-Net and DilatedNet.Discussion and conclusion
This study represents the first effort to synthesize quantitative CBF from BOLD fMRI. the proposed BOA-Net produced CBF quantification results with higher SNR, higher temporal resolution, both were inherited from BOLD fMRI (higher SNR is also contributed by DL denoising). For the existing dataset without ASL MRI acquired, BOA-Net offers a unique tool to generate a new functional imaging modality. For data with ASL MRI acquired, the BOA-Net derived CBF data can provide a re-test dataset for the CBF data measured by ASL MRI.Acknowledgements
This work was supported by NIH/NIA grant: 1 R01 AG060054-01A1References
[1] Lehtinen, Jaakko, et al. "Noise2Noise: Learning Image Restoration without Clean Data." International Conference on Machine Learning. 2018. [2] Kim, Ki Hwan, Seung Hong Choi, and Sung-Hong Park. "Improving Arterial Spin Labeling by using deep learning." Radiology 287.2 (2017): 658-666. [3] Yu, Jiahui, et al. "Wide activation for efficient and accurate image super-resolution." arXiv preprint arXiv:1808.08718 (2018). [4] Shin, David D., et al. "Pseudocontinuous arterial spin labeling with optimized tagging efficiency." Magnetic resonance in medicine 68.4 (2012): 1135-1144. [5] Li, Yiran, et al. "Priors-guided slice-wise adaptive outlier cleaning for arterial spin labeling perfusion MRI." Journal of neuroscience methods 307 (2018): 248-253. [6] Wang, Ze, et al. "Empirical optimization of ASL data analysis using an ASL data processing toolbox: ASLtbx." Magnetic resonance imaging 26.2 (2008): 261-269. [7] Ogawa, Seiji, et al. "Brain magnetic resonance imaging with contrast dependent on blood oxygenation." proceedings of the National Academy of Sciences 87.24 (1990): 9868-9872. [8] Detre, John A., et al. "Perfusion imaging." Magnetic resonance in medicine 23.1 (1992): 37-45. [9] Ronneberger, Olaf, Philipp Fischer, and Thomas Brox. "U-net: Convolutional networks for biomedical image segmentation." International Conference on Medical image computing and computer-assisted intervention. Springer, Cham, 2015. [10] F. Yu and V. Koltun, "Multi-scale context aggregation by dilated convolutions," arXiv preprint arXiv:1511.07122, 2015.Figures
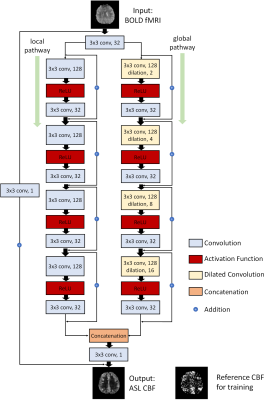
Figure 1: Illustration of the architecture of DWAN. The
output of the first layer was fed to both local pathway and global pathway.
Each pathway contains 4 consecutive wide activation residual blocks. Each wide
activation residual block contains two convolutional layers (3×3x128 and
3×3x32) and one activation function layer. The 3x3x128 convolutional layers in
the global pathway were dilated convolutional layers [10] with a dilation rate
of 2, 4, 8, 16, respectively (a×b×c indicates the property of convolution.
a×b is the kernel size of one filter and c is the number of the filters).
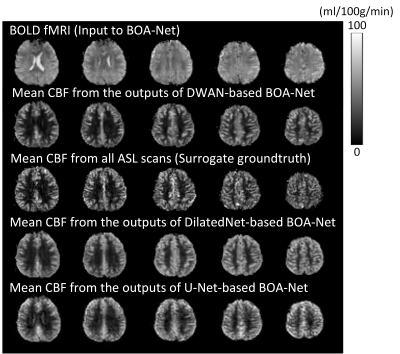
Figure 2.
BOLD fMRI and CBF images from one representative subject. Only 5 axial slices
were shown. From top to bottom: slice 7, 8, 9, 10, 11.
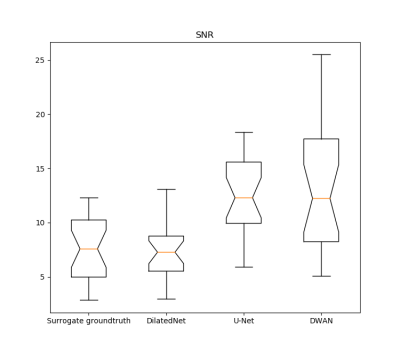
Figure
3. The notched box plot of the SNR from surrogate groundtruth and mean
CBF maps from the outputs of BOA-Net with different network architectures. SNR was calculated from 23 test subjects.