3887
To smooth or not to smooth? Modelling the impact of spatially correlated physiological noise in fMRI1Wellcome Centre for Human Neuroimaging, UCL Institute of Neurology, London, United Kingdom
Synopsis
Spatial smoothing is common in fMRI analyses but the benefit to functional sensitivity can vary depending on baseline signal-to-noise, inherent smoothness and physiological noise characteristics. Here we propose an extended model that can quantify each of these properties in addition to parameterising the degree of spatial correlation in the physiological noise. The model is validated through simulation and in vivo experiment. This new model allows the complete characterisation of the impact of spatial smoothing from a single fMRI time series enabling researchers to efficiently gain insight into their data and to optimise processing pipelines.
Introduction
Spatial smoothing is common when post-processing fMRI time-series. The impact on the temporal SNR (tSNR) depends on the baseline SNR, the intrinsic smoothness of the data (σI) and the characteristics of the physiological noise, including both amplitude (λ) and the degree of spatial correlation1. Here we propose an extended model that additionally parameterises the degree of spatial correlation of the physiological noise (α). Furthermore, this model can be used to estimate σI, λ and α from just one time-series. We validate the model with simulated and in vivo 3T data. We replicate the finding that physiological noise increases with the number of segments with 3D-EPI2.Methods: Model extension
Consider a time series comprised of a mean signal $$$\bar{S}$$$ with variance components σ02 and σp2 arising from thermal and physiological noise processes respectively, with the physiological component varying with signal intensity3 (σp=λS).Images have some intrinsic smoothness due to the acquisition and reconstruction processes. Applying an isotropic 3D Gaussian filter to a volume with spatially uncorrelated noise reduces the variance by $$$8\pi^{3/2}\sigma_F^3$$$ where σF is the standard deviation (SD) of the filter. Therefore, by approximating the intrinsic smoothness with such a filter, with SD=σI, the baseline SNR of the original images can be expressed as: $$$SNR_o=\frac{\bar{S}}{\sigma_0}\sqrt{8\pi^{3/2}\sigma_I^3}$$$.
Similarly, spatial smoothing with a filter with SD=σF decreases the noise variance without affecting the mean signal such that:$$$\left(\frac{SNR_{smooth}}{SNR_o}\right)^2=\frac{\sqrt{\sigma_I^2+\sigma_F^2}^3}{\sigma_I^3}=V\space\space(Eq.1)$$$. This ratio is denoted V for consistency with1. We introduce an additional parameter α tuning the degree of spatial correlation in the physiological noise such that: $$$\sigma_{p_{smooth}}=\frac{\sigma_p}{1+\alpha\left(\sqrt{8\pi^{3/2}\sqrt{\sigma_I^2+\sigma_F^2}^3}-1\right)}$$$ . In the extrema, if α=0, the physiological noise is fully spatially correlated, such that smoothing does not reduce the variance, whereas if α=1, the physiological noise is spatially uncorrelated and smoothing reduces the variance in a manner equivalent to smoothing the thermal noise.
The tSNR of the smoothed time-series can therefore be expressed as:
$$tSNR_{smooth}=\frac{\bar{S}}{\sqrt{\sigma_{0_{smooth}}^2+\sigma_{p_{smooth}}^2}}=\frac{V.SNR_o}{\sqrt{V+V^2\lambda^2SNR_o^2\left(1+\alpha\left(\sqrt{V8\pi^{3/2}\sigma_I^3}-1\right)\right)^{-2}}}\space \space (Eq.2)$$
Methods: Validation
- Simulated data
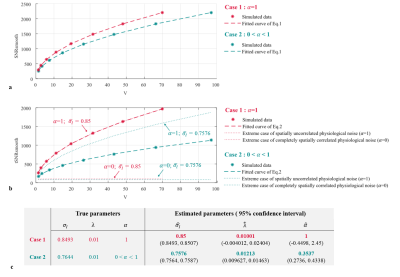
A second set of data were created as above, but with the additional factor of spatially correlated physiological noise, $$$\mathcal{N}(0,\sigma_p^2C)$$$ , added in the first step. tSNRsmooth was computed from these smoothed time-series. SNRsmooth,SNRo and tSNRsmooth were then averaged across the centre portion of the grid (to avoid edge effects). The ratio $$$\left(\frac{SNR_{smooth}}{SNR_o}\right)^2 $$$ was fit to Eq.1 in order to estimate σI, which was subsequently used to fit tSNRsmooth to Eq. 2 to estimate α and λ.
Two cases were investigated. Physiological noise was either:
1./ Spatially uncorrelated: $$$C=I_d$$$
2./ Partially spatially correlated: C was a symmetric positive definite matrix with some off-diagonal elements non-null.
- In vivo data
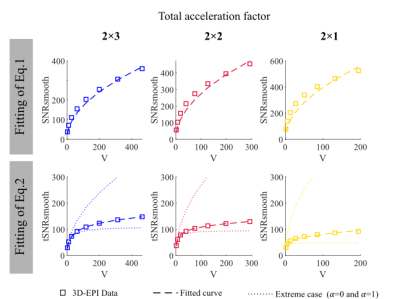
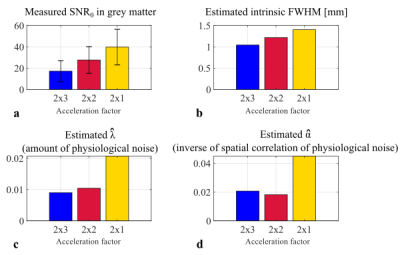
SNR was computed using a Monte-Carlo method6. A set of replica volumes were reconstructed from the time-series’ mean k-space data with additive noise sampled from a distribution with the same statistics as the thermal noise obtained from the same protocols with no RF excitation. The replicas and the acquired volumes were smoothed with a 3D Gaussian filter with FWHM from 1 to 8 mm. SNRo and SNRsmooth were computed as the ratio of the mean and the SD across replicas before and after smoothing, respectively. tSNRsmooth was computed in the same way from the acquired smoothed time-series after additional realignment and detrending. tSNRsmooth, SNRo and SNRsmooth were averaged across grey matter (p(GM)>0.99) after correcting for non-Gaussianity7. The summary data were fit to (Eq.1) and (Eq.2).
Results
- Simulated data
- In vivo data
Fig.2 and Fig.3b-c-d show results of the model fitting. The estimated intrinsic smoothness followed the same pattern as the SNR. $$$\hat{\lambda}$$$ increased with the number of acquired partitions (from 0.009 to 0.0207), consistent with previous findings2. $$$\hat{\alpha}$$$ was neither 0 nor 1 indicative of partially correlated physiological noise, as previously observed in1.
Discussion
The extended model proposed here allows the full impact of smoothing to be characterised without the need to acquire several time series with variable baseline SNR to estimate the amplitude of the physiological noise(λ). The model results, validated via simulation and in vivo experiment, are consistent with literature.Acknowledgements
The WCHN is supported by core funding from the Wellcome [203147/Z/16/Z].References
1. Triantafyllou C, Hoge RD, Wald LL. Effect of spatial smoothing on physiological noise in high-resolution fMRI. NeuroImage 2006;32:551–557 doi: 10.1016/j.neuroimage.2006.04.182.
2. Zwaag W van der, Marques JP, Kober T, Glover G, Gruetter R, Krueger G. Temporal SNR characteristics in segmented 3D-EPI at 7T. Magnetic Resonance in Medicine 2012;67:344–352 doi: 10.1002/mrm.23007.
3. Krüger G, Glover GH. Physiological noise in oxygenation-sensitive magnetic resonance imaging. Magn Reson Med 2001;46:631–637 doi: 10.1002/mrm.1240.
4. Lutti A, Thomas DL, Hutton C, Weiskopf N. High-resolution functional MRI at 3 T: 3D/2D echo-planar imaging with optimized physiological noise correction. Magn Reson Med 2013;69:1657–1664 doi: 10.1002/mrm.24398.
5. Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: Sensitivity encoding for fast MRI. Magnetic Resonance in Medicine 1999;42:952–962 doi: 10.1002/(SICI)1522-2594(199911)42:5<952::AID-MRM16>3.0.CO;2-S.
6. Robson PM, Grant AK, Madhuranthakam AJ, Lattanzi R, Sodickson DK, McKenzie CA. Comprehensive Quantification of Signal-to-Noise Ratio and g-Factor for Image-Based and k-Space-Based Parallel Imaging Reconstructions. Magn Reson Med 2008;60:895–907 doi: 10.1002/mrm.21728.
7. Constantinides CD, Atalar E, McVeigh ER. Signal-to-noise measurements in magnitude images from NMR phased arrays. Magnetic Resonance in Medicine 1997;38:852–857 doi: 10.1002/mrm.1910380524.
Figures