3810
A joint-community effort to standardize quantitative MRI data: Updates from the BIDS extension proposal1NeuroPoly Lab, Institute of Biomedical Engineering, Polytechnique Montreal, Montreal, QC, Canada, 2Montreal Heart Institute, Montreal, QC, Canada, 3Laboratory for Social and Neural Systems Research (SNS Lab), Department of Economics, University of Zurich, Zürich, Switzerland, 4Spinoza Centre for Neuroimaging, Amsterdam, Netherlands, 5Center for Adaptive Rationality, Max Planck Institute for Human Development, Berlin, Germany, 6University of Surrey, Guildford, United Kingdom, 7Stanford University, Stanford, CA, United States, 8Department of Medical Biophysics, Robarts Research Institute, University of Western Ontario, London, ON, Canada, 9Wellcome Centre for Integrative Neuroimaging, FMRIB, Nuffield Department of Clinical Neurosciences, University of Oxford, Oxford, United Kingdom, 10GIGA Institute, University of Liège, Liège, Belgium, 11Department of Psychiatry, University of Cambridge, Cambridge, United Kingdom, 12The Alan Turing Institute, London, United Kingdom
Synopsis
The Brain Imaging Data Structure (BIDS) aims to develop a standard for organizing and describing neuroimaging data (https://bids.neuroimaging.io/). It has rapidly gained traction across neuroimaging disciplines through community-driven development of BIDS extension proposals (BEPs; http://bit.ly/bids_bep). Here were present such an extension proposal, that captures a wide range of structural MRI contrasts and parametric mapping protocols that are of interest to the broader ISMRM community (https:/bit.ly/bep001). This abstract reports the current state of the proposal and illustrates the developments made since the 2018 ISMRM virtual meeting Bringing BIDS closer to quantitative MRI.
INTRODUCTION
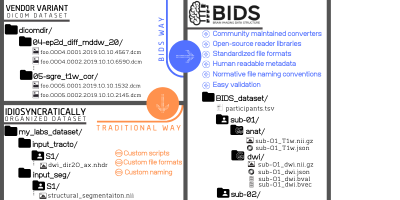
As neuroimaging research is becoming increasingly data-intensive and with the surgeance of dataset sharing within the last decade, proper data standardization and annotation is of paramount importance1. The Brain Imaging Data Structure2,3 (BIDS) aims to develop a standard for organizing and describing neuroimaging data.Different vendors and labs have different conventions for organizing neuroimaging data, which makes it difficult to standardize processing (Figure 1). BIDS standardizes this organisation, which eases the development of open-source turn-key BIDS apps and sharing of public datasets. Its applicability has steadily been increased by the community-driven development of BIDS extension proposals (BEP).
Over the last two years, we have worked to bring MRI physicists and neuroimaging researchers together and collaborate on a BIDS specification to facilitate the standardization and use of quantitative MRI (qMRI) methods by neuroscientists. In this work, we introduce our specification, share example datasets, and illustrate how multiple BIDS apps4 can easily work on the same BIDS-compliant qMRI data.
METHODS
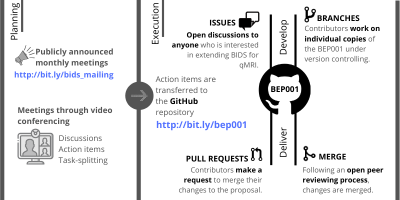
Community-driven open developmentBEP001 contributors have been holding monthly video-conferences over the last two years, making consensus decisions for improvement and sharing action items. The standard operational procedure to advance our proposal is summarized in Figure 2.
qMRI within the BIDS framework
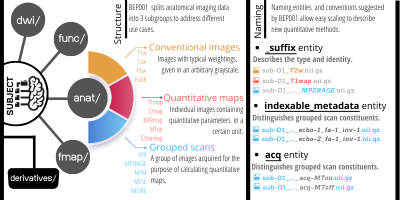
BIDS was originally developed by functional neuroimaging researchers who laid out a directory structure covering anatomical, functional, diffusion-weighted and field map images. BEP001 focuses on structural (anatomical) acquisitions that include multiple contrasts and sensitivity maps. Therefore has placed all qMRI data in the anat and fmap folders.
In the BIDS specification the suffix of the file name indicates the type of data captured in the image, for example _T1w and _bold. In this extension, we added 3 categories of suffixes to annotate the use-case of more advanced anatomical imaging data: i) conventional images, ii) quantitative maps and iii) grouped scan collections (Figure 3). Grouped scan collections identify protocols where multiple images are acquired with largely identical parameters, except for a few key parameters such as inversion time, echo time or flip angle.
We introduce the indexable_metadata-<index> entity to capture such key parameters. For example a multi-echo gradient echo-recalled sequence will include images with the key pairs _echo-1 and _echo-2 in their respective filenames. If the parameters that vary between the constituents of a grouped scan are categorical rather than numerical, the _acq-<label> entity is used. BEP001 suggests a list of labels to be used with the _acq key tag for the sake of creating a coherent naming entity for grouped scans. Currently available _acq labels are MTon, MToff, and T1w, creating sensible filenames for e.g., magnetization transfer saturation6 (MTsat) and multi-parameter mapping7 .
BIDS makes a strong distinction between "raw" images that are directly imported from the scanner and "derived" images that have been processed in some way. However, it is important to emphasize that "raw" in the BIDS context does not refer to MR k-space data before reconstruction, which means that BIDS is compatible with the ISMRM raw data format8. To maintain consistency and ensure compatibility between the two standards, we propose that quantitative parameter maps that are estimated with a model are stored in the /derivatives folder, whereas unprocessed are stored in the raw /anat folder. To aid BIDS processing pipelines that only take raw data as an input, researchers can use file links to make parameter maps also available in the raw data folder.
Metadata for grouped scan collections
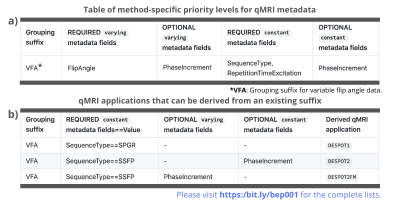
In addition to a file naming system, BIDS also prescribed how to store relevant metadata in a "sidecar" json-file. This is especially relevant for qMRI applications, where key imaging parameters are essential for further processing. Our proposal standardizes which parameters are required and optional for specific grouped acquisition protocols (e.g., MP2RAGE, VFA; Figure 4a). It also standardizes optional parameters that are relevant for specific qMRI applications (Figure 4b).
RESULTS
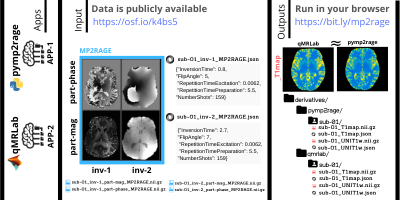
We created an open data repository9 and uploaded example qMRI datasets adhering to the BEP001 proposal. To demonstrate a use case, we used two open-source software packages, pymp2rage10 and qMRLab11,12, for T1 mapping using MP2RAGE data from our open repository (Figure 5). The analyses are version controlled on GitHub and fully reproducible, with no installation, at https://bit.ly/mp2rage.So far, BEP001 has merged 22 pull requests and hosted 48 issues with the contributions of more than 10 developers around the globe. Currently it lists 5 descriptions for conventional contrasts, 14 for qMRI maps and 9 for grouped scan collections along with their metadata priority level definitions. A sizable multi-center public database for spinal cord MRI13, which attracted tens of thousands of viewers in a few months, has already adopted conventions suggested by BEP001. Members of the ISMRM community are invited to join the development process to make their methods easy to implement for researchers in the cognitive and clinical neurosciences.
CONCLUSION
In this work, we introduced the core components of BEP001, shared real-world example datasets and demonstrated how applications written in different languages can easily work on the same BIDS compliant qMRI data. We hope that these efforts will not only better organize our data, but will also facilitate multi-site collaborative work and encourage neuroscientists to adopt advanced MR techniques.Acknowledgements
The authors A.K. and G.H. contributed equally to this work.
References
[1] Poldrack, R., et al. “Scanning the horizon: towards transparent and reproducible neuroimaging research.” Nature Reviews Neuroscience 18 (2017): 115-126.
[2] Gorgolewski, K., et al. "The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments." Scientific Data 3 (2016): 160044.
[3] https://bids.neuroimaging.io/, Accessed November 5, 2019.
[4] Gorgolewski, K., et al. "BIDS apps: Improving ease of use, accessibility, and reproducibility of neuroimaging data analysis methods." PLoS Comp. Biol. 13.3 (2017): e1005209.
[5] https://github.com/bids-standard/bep001, Accessed November 5, 2019.
[6] Helms, G., et al. "High‐resolution maps of magnetization transfer with inherent correction for RF inhomogeneity and T1 relaxation obtained from 3D FLASH MRI." Magn. Reson. Med. 60 (2008): 1396-1407.
[7] Weiskopf, N., et al. "Quantitative multi-parameter mapping of R1, PD*, MT, and R2* at 3T: a multi-center validation." Frontiers in Neuroscience 7 (2013): 95.
[8] Inati, S., et al. “ISMRM Raw data format: A proposed standard for MRI raw datasets”, Magn. Reson. Med. 7 (2017): 411-421.
[9] Karakuzu, A., et al. (2019). BEP[001]. https://doi.org/10.17605/OSF.IO/K4BS5, Accessed October 28, 2019.
[10] https://github.com/Gilles86/pymp2rage, Accessed November 5, 2019.
[11] Duval, T., et al. “Quantitative MRI made easy with qMRLab”, International Society for Magnetic Resonance in Medicine Meeting (2018): Abstract 2288.
[12] https://qmrlab.org, Accessed November 5, 2019.
[13] Cohen-Adad, J., et al. “Spinal Cord MRI Public Database (multi-subjects)”, https://openneuro.org/datasets/ds001919/versions/1.0.4, DOI: 10.18112/openneuro.ds001919.v1.0.4, Accessed October 26, 2019.
Figures