3687
Connectivity-based Graph Convolutional Network for fMRI Data Analysis
Lebo Wang1, Kaiming Li2, and Xiaoping Hu1,2
1Department of Electrical and Computer Engineering, University of California, Riverside, Riverside, CA, United States, 2Department of Bioengineering, University of California, Riverside, Riverside, CA, United States
1Department of Electrical and Computer Engineering, University of California, Riverside, Riverside, CA, United States, 2Department of Bioengineering, University of California, Riverside, Riverside, CA, United States
Synopsis
Graphs have been widely applied for ROI-based fMRI data analysis, in which the functional connectivity (FC) between all pairs of regions is thoroughly considered. Combined with convolutional neural networks, we define graphs based on FC and introduce a connectivity-based graph convolution network (cGCN) architecture for fMRI data analysis. cGCN allows us to extract spatial features within connectivity-based neighborhood for each frame and capture the temporal dynamics between frames. Our results indicate that cGCN outperforms traditional deep learning architectures on fMRI data analysis.
INTRODUCTION
There has been success with deep learning in fMRI analysis, where fMRI data are considered on structured grids and spatial features within Euclidean neighbors are extracted through convolutional neural networks (CNNs). Meanwhile, graphs, as a ubiquitous data structure in many applications, have been widely applied for ROI-based fMRI data analysis, in which functional connectivity (FC) between all pairs of regions is thoroughly considered. Inspired by the remarkable performance of recently introduced CNNs on graph data, we define graphs based on FC and introduce a connectivity-based graph convolution network (cGCN) architecture for fMRI data analysis. This architecture allows us to extract spatial features within connectivity-based neighborhood from each frame and capture the temporal dynamics of fMRI data.METHODS
We considered fMRI data analysis as a spatiotemporal feature extraction task, where spatial patterns were extracted from each frame, and temporal evolution was obtained between frames based on existing spatial features. For each frame of fMRI data, ROI-based fMRI data were represented in the form of graphs, in which ROIs acted as nodes with intensity values and the edge weight between any two ROIs was given by their Pearson’s correlation coefficient. In order to reduce the computational complexity, a k-nearest neighbor (k-NN) graph was defined by taking the top-k neighbors for each node with the highest correlation. In this way, the community information was also reserved in the k-NN graph. Different values (3, 5, 10 and 20) of k, the only hyperparameter related to the graph structure and similar to the size of the convolution kernel, were tested in our study. Stacking convolutional layers generated spatial features with the shared k-NN graph, and latent representations were obtained by a recurrent neural network (RNN) and followed by a softmax layer for classification. The architecture of cGCN is shown in Fig.1.We trained and tested our cGCN architecture to realize individual identification on 100 subjects with resting-state fMRI data from the Human Connectome Project (54 females, mean age = 29.1 ± 3.7, TR = 0.72 s)1. Two scans on the 1st day were used for training and other two scans on the 2nd day were used for validation and testing. Each scan has 1200 frames in total. During training and validation, input data were divided into 100-frame segments. The final identification accuracy was reported as the average performance on the testing dataset with different number of frames as inputs. In order to validate the contribution of connectivity-based neighborhood for convolutions, we compared the cGCN performance with the FC-based k-NN graph and a random graph.
RESULTS
In Fig.2, we show the individual identification performance on the testing dataset with different number of frames (from 1200 frames to a single frame) as input data. In general, cGCN achieved better performance with increasing number of frames and its performance leveled off with over 100 frames. In terms of the k value, cGCN achieved highest identification accuracy on average when k=10, compared with other k values (3, 5, and 20). Compared to the traditional CNN model2, cGCN showed substantial performance improvement with fewer than 100 frames. For cGCN models with the random k-NN graph, the classification accuracy was always lower than cGCNs, however, they still outperformed the conventional CNN significantly, especially when k=3 and 5.DISCUSSION
fMRI data can be represented on graphs with top connectivity-based neighborhood being directly connected. Instead of considering fMRI data as grid data and performing convolutions within neighbors on the grid in Euclidean space, we conducted convolutions within top connectomic neighbors for spatial features. In such a way, the spatial pattern related to FC can be extracted by stacking convolutional layers within multi-hop neighborhood for the enlarged receptive field. Our cGCN achieved better individual identification than traditional CNNs, which performed convolution on the list of ROIs without considering appropriate neighborhood. Our results also demonstrated that cGCN model with k=10 achieved the highest identification accuracy. One possible reason is that increasing the number of convolutional neighbors can utilize more connectomic neighbors for complex feature extraction. However, convolutions on too many nodes could fail to generate local generalizable features. For this application, the k value of 10 is a good compromise between these opposing factors, although the optimal value of k will depend on the application.CONCLUSION
We introduced a graph convolutional neural network with neighborhood defined based on FC, cGCN, for resting-state fMRI data analysis. Our cGCN was evaluated on HCP data with varying number of neighborhoods. The experimental results indicate that 1) the new architecture outperforms existing approaches and 2) a neighborhood size of 10 is optimal for individual identification. The new approach provides a robust alternative for analyzing resting-state fMRI data.Acknowledgements
Thanks to Alexandra Reardon for proofreading.References
- Van Essen D C, Smith S M, Barch D M, et al. The WU-Minn Human Connectome Project: An Overview. Neuroimage. 2013. 80: 62-79.
- Wang L, Li K, Chen X, et al. Application of Convolutional Recurrent Neural Network for Individual Recognition Based on Resting State Fmri Data. Frontiers in Neuroscience. 2019.
Figures
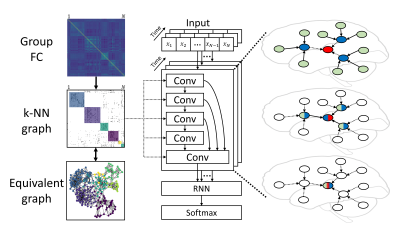
Fig.1. Overview of the cGCN
architecture for fMRI data. Group FC was obtained and the k-NN graph was built
to keep top-correlative neighborhood for each node. Spatial features were
extracted by convolutional layers and temporal dynamics were modeled by the RNN
layer. The final latent representations were classified by the softmax layer.
The increasing receptive
field is visually illustrated on the right. The central node is colored in red,
and is connected by direct neighborhoods in blue and 2-hop neighbors in green.
Spatial features were extracted after two convolutional layers.

Fig.2. Individual identification
performance on the HCP dataset. The classification accuracy of cGCN is shown
with different number of frames as inputs. The highest classification accuracy
on average was achieved when k=10. However, the accuracy dropped when k reached
20. The performance improvement over cGCN with random graphs proved that
connectivity-based neighborhood is vital for spatial feature extraction.
Compared to the traditional CNN model, cGCN showed great performance
improvement with less than 100 frames as input data.