3669
Quantification of brain age using high-resolution 7T MR imaging and implications in major depressive disorder1Radiology, Icahn School of Medicine at Mount Sinai, NEW YORK, NY, United States, 2Psychiatry, Icahn School of Medicine at Mount Sinai, NEW YORK, NY, United States
Synopsis
A regression model estimating brain-age from about 250 T1-weighted imaging features was developed using data from 29 healthy controls (mean age 39.8). The model estimated brain age with average absolute error of 6.0 years. The model was applied 35 patients (mean age 38.7) diagnosed with major depressive disorder (MDD), yielding similar performance (7.6 years mean absolute error), but showed trend of over-estimation of average brain-age by 2.4 years. This technique demonstrates the feasibility of brain-age estimation using imaging features, and may help assess the differential effects of pathology like MDD in the aging process.
Introduction
Aging manifests in multiple ways in human brain morphology, including atrophy of gray matter and white matter, thinning of cortical volumes and a consequential increase in cerebrospinal fluid volume in the ventricles. Brain morphometry has generated extensive interest in major depression, with particular focus in the hippocampus, but also in cortical regions. Previous studies have also suggested that these volumetric effects may dependent on age or disease duration, suggesting a need for modeling healthy and pathological aging in the brain through imaging.With the benefits of high resolution provided by ultrahigh-field, anatomical T1-weighted MR may be combined with automatic segmentation and quantification techniques like Freesurfer to generate imaging parameters precisely describing the brain morphology. Modeling these image parameters may yield markers for brain-age, and potentially identify the effects of psychiatric disorders like major depressive disorder on the brain aging process.
In this study, a linear regression model weighted for age-correlation was developed using brain morphology data from a cohort of healthy volunteers. The model was then applied on a separate cohort of patients diagnosed with MDD to probe the differential effects of MDD on the age relationship of these imaging markers.
Materials & Methods
Thirty-five patients diagnosed with major depressive disorder (MDD) (14 female, 21 male, age 38.7±11.0 years) and twenty-nine healthy controls (9 female, 20 male, age 39.8±10.4 years) were scanned on 7T MRI (Siemens Magnetom, 32Rx/1Tx channel Nova head coil) using a T1-weighted MPRAGE sequence and the following parameters: TE/TR: 3.62,s/6000ms, 224x168 mm3 FOV, 320x240 array size, 240 slices, 0.7 mm3 isotropic resolution, 7:26 minute acquisition time. All scan data was subsequently processed using Freesurfer version 6.0 to perform automatic segmentation and cortical & subcortical voxel-based volumetrics. These imaging parameters were collected into four categories: whole brain measures like white/gray matter volume and ratios, cortical thickness, cortical gray/white ratios, normalized cortical volume and normalized subcortical volumes. In all, 258 imaging features were selected, representing a subset of the features considered in a previous brain-age study of the UK BioBank. Single linear regressions were performed on each feature, and separately for male and female subjects, and two sets of slopes, intercepts and p-values were computed in Matlab. The inverse-square of the p-values was used to weight each feature to generate a model to estimate brain age. The computed model was subsequently applied to analogously-processed T1-imaging data features from MDD patients.Results
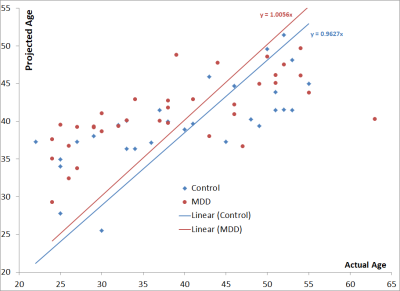
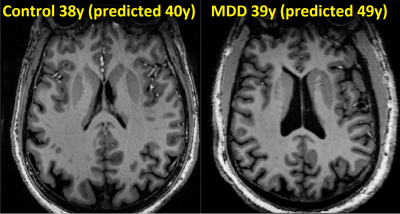
The regression model showed an average absolute error of 6 years in among the control subjects, which in this cohort represents an error rate of 15%. When applied to a separate cohort of MDD patients the model produced a similar error rate of 7.6 years (or 19%), and the slight discrepancy in performance may be explained by 2.4 year over-estimation in the age of the MDD patients (41.1 years predicted vs. 38.7 years average actual age). A student’s t-test comparing the mean signed error between patients and controls showed no significant difference between patients and controls with a p value of 0.240. Figure 1 shows a scatterplot showing the relationship between actual age and predicted age among the controls and MDD subjects.To reduce the potential for over-fitting the control cohort, the model was generated with simple linear regressions and weighted by the squared inverse of their p-values. This weighting would de-emphasize contributions from imaging features whose relationship with aging is not strongly linear. Among the imaging features that showed the greatest correlation with age (and thereby carrying the greatest weight in the model) were the whole-brain gray matter volumes, total cortical, normalized precentral, parietal and middle temporal lobe volumes as well as gray/white matter ratios of the precuneus and pars triangularis. Figure 2 shows an appropriately-classified control subject and an MDD patient whose age was over-estimated by based on these criteria.
The model consisting of separate regressions for male and female subjects produced the lowest total error, though similar performance and MDD age gaps were observed when assessing models which either didn’t consider gender (6.9 year average absolute error, 2.7 year MDD age over-estimation) or regressed it out (6.7 year average absolute error, 2.4 year MDD age over-estimation). The lack of significant difference between the age over-estimation of patients and controls may reflect imprecision in the model or the limited number of quantified subjects. A non-linear regression or machine learning based approach may yield more precise estimation of brain age, though over-fitting may remain a concern in this limited dataset. Because imaging parameters are based on morphological features rather than radiomics/contrast derived features, it may be possible to expand the dataset using data from other cohorts, including those scanned at lower field strengths.
Conclusion
Brain-age models based on imaging features show the potential to estimate the morphological effects of aging. Refining these models with more sophisticated fitting and larger datasets may facilitate their ability to probe the effects of psychiatric disorders like MDD on brain aging.Acknowledgements
The authors would like to acknowledge funding from NIH R01 MH109544.References
1. Lange, C. and E. Irle, Enlarged amygdala volume and reduced hippocampal volume in young women with major depression. Psychological medicine, 2004. 34(6): p. 1059-1064.
2. Bremner, J.D., et al., Reduced volume of orbitofrontal cortex in major depression. Biological psychiatry, 2002. 51(4): p. 273-279.
3. Steffens, D.C., et al., Hippocampal volume in geriatric depression. Biological psychiatry, 2000. 48(4): p. 301-309.
4. Miller, K.L., et al., Multimodal population brain imaging in the UK Biobank prospective epidemiological study. Nature neuroscience, 2016. 19(11): p. 1523.
5. Cole, J.H., et al., Predicting brain age with deep learning from raw imaging data results in a reliable and heritable biomarker. NeuroImage, 2017. 163: p. 115-124.
6. Franke, K., et al., Estimating the age of healthy subjects from T1-weighted MRI scans using kernel methods: exploring the influence of various parameters. Neuroimage, 2010. 50(3): p. 883-892.
Figures