3654
Development of a management system for radiology–common data model (R-CDM) and its application in the liver disease: extension of OMOP-CDM
Min-Gi Pak1, Seong-Min Han2, ChungSub Lee3, SeungJin Kim1, Tae-Hoon Kim3, Chang-Won Jeong3, and Kwon-Ha Yoon3,4
1Medical Science, Wonkwang University, Iksan, Republic of Korea, 2Computer Software Engineering, Wonkwang University, Iksan, Republic of Korea, 3Medical Convergence Research Center, Wonkwang University, Iksan, Republic of Korea, 4Radiology, Wonkwang University, Iksan, Republic of Korea
1Medical Science, Wonkwang University, Iksan, Republic of Korea, 2Computer Software Engineering, Wonkwang University, Iksan, Republic of Korea, 3Medical Convergence Research Center, Wonkwang University, Iksan, Republic of Korea, 4Radiology, Wonkwang University, Iksan, Republic of Korea
Synopsis
The Observational Medical Outcomes Partnership-Common Data Model (OMOP-CDM) used in distributed research networks has low coverage of clinical data and does not reflect the latest trends of precision medicine. Radiology data have great merits to visual and identify the lesions in specific diseases. However, radiology data should be shared to obtain the sufficient scale and diversity required to provide strong evidence for improving patient care. Our study was to develop a web-based management system for radiology-CDM (R-CDM), as an extension of the OMOP-CDM, and to evaluate the feasibility of R-CDM dataset for application of radiological image data in clinical practice.
Introduction
To date, the distributed research network has been adopted by global research collaboration groups, including the Observational Health Data Sciences and Informatics (OHDSI) consortium. The Observational Medical Outcomes Partnership-Common Data Model (OMOP-CDM) was developed by the OHDSI consortium and includes clinical data of electronic health records (EHR) from over 20 countries, with information of 1.5 billion patients transformed to date. However, OMOP-CDM used in distributed research networks has low coverage of clinical data and does not reflect the latest trends of precision medicine.Recently, a research group belong to OHDSI developed genomic CDM (G-CDM), as an extension of the OMOP-CDM to improve clinical data coverage. G-CDM provided the effective integration of genomic data with standardized clinical data, allowing for data sharing across institutes. Compared to EHR (or EMR) and genomic data, radiological image data have great merits to visual and identify the lesions in specific diseases. However, radiology data should be shared in order to achieve the sufficient scale and diversity required to provide strong evidence for improving patient’s diagnosis and care. Thus, a distributed research network for radiology data allows researchers to share this evidence rather than the patient-level data across centers, thereby avoiding privacy issues.
Therefore, the aim of this study was to develop a web-based management system for radiology-CDM (OHDSI proposed R-CDM), as an extension of the OMOP-CDM, and to evaluate the feasibility of R-CDM dataset for application of radiological image data in clinical practice.
Methods
Data structure of the Radiology-Common Data Model (R-CDM)Data structure for R-CDM is basically used the OMOP-CDM structure (Fig. 1). To link clinical data in the OMOP-CDM (Condition_Occurrence, blue box), the following information on each patient with radiological image data was stored in a separate corresponding table: Radiology_Occurrence, Radiology_Image, Radiology_Protocol, Radiology_Modality, Radiology_Device, and Radiology_Hospital, respectively (Fig. 2).
Standardization of terminology for R-CDM
Terminology of OMOP-CDM is used “SNOMED” and “SNOMED Clinical Terms® (SNOMED CT®)” for standardization of terminology. SNOMED and SNOMED CT® was originally created by the College of American Pathologists. “SNOMED”, “SNOMED CT” and “SNOMED Clinical Terms” are registered trademarks of the SNOMED International (www.snomed.org). Also, a web service of standardized vocabularies (called ‘Athena’) is available at http://athena.ohdsi.org/search-terms/terms (Fig. 3). In order to standardize the R-CDM vocabulary, R-CDM data are used not only “SNOMED” as OMOP-CDM, but also “RadLex radiology lexicon” produced from Radiological Society of North America (RSNA), available at https://www.rsna.org/en/practice-tools/data-tools-and-standards/radlex-radiology-lexicon.
Management system of R-CDM
Management system of R-CDM developed by web-based client server architecture using Python-Django Rest Framework and JavaScript language-based React library. The dataset standardization procedure was as follows: selection of clinical condition, uploading radiological image dataset, extraction of metadata, and build standard R-CDM dataset. The system provided searching & downloading functions, Occurrence List Viewer and Image Viewer.
Data description of chronic liver disease for clinical application
For the construction of a R-CDM dataset, the study design was retrospective study and the study protocol was approved by the institutional review board (IRB) of our University Hospital. A total of 1637 patients with suspected chronic liver disease (CLD) were recruited from January 2002 to December 2018. This study standardized a CLD R-CDM dataset consisting of MRI (n=111) and CT data (n=1526). The disease code for chronic liver disease is obtained in SNOMED Concept Code (328383001) (Fig. 3). Also, the private information such as Patient Name (DICOM header Tag No.= 0010, 0010), Patient ID (0010, 0020), Patient Sex (0010, 0040), and Patient Age (0010, 1010) are deleted for data anonymization to prevent the identification of the of patient (see Table 1). The quality of final CLD R-CDM dataset was evaluated five domains by four expert radiologists (with more than 10 years of experience) and four fellows. The domains are consisted of dataset composition, patient selection, standard terminology, detailed data quality (completeness/validity/accuracy/uniqueness/consistency) and data anonymization (each domain min. 10 – max. 100; total 500).
Results & Discussion
For a distributed research network and easy multicentric study, we developed a web-based management system for R-CDM. Also, we constructed a clinical R-CDM dataset as CLD dataset by standardizing 145,188 MR images (n=111) and 620,389 CT images (n=1526). The averaged uploading time for dataset was CT 40.2±2.0 sec (per 150 images) and MRI 43.6±16.2 sec (per 150 images), and the averaged conversion time for standardization was CT 44.8±31.5 sec (per 150 images) and MRI 28.0±11.9 sec (per 150 images). In the dataset quality, averaged scores in five domains are dataset composition 81±16, patient selection 82±5, standard terminology 81±22, detailed data quality 83±10 and data anonymization 92±8 (Total score= 419±40). Figure 4 showed the representative standardized data on the web-based management system with Occurrence Viewer. Our system allowed the standardization code of the SNOMED vocabulary and RadLex Term to search and download dataset using keywords. In addition, the management system provided the Image Viewer for showing the detail information (Fig. 4).Conclusion
This study proposed a radiology–common data model (R-CDM) in conjunction with OMOP-CDM for a distributed research network. We developed a web-based management system for searching and downloading standardized R-CDM dataset and constructed a chronic liver disease R-CDM dataset. Our management system and CLD dataset would be useful for multicenter study and machine learning research.Acknowledgements
This study was supported by the Korea Health Technology R&D Project through the Korea Health Industry Development Institute(KHIDI), funded by the Ministry of Health & Welfare(HI18C1216) and the Technology Innovation Program (or Industrial Strategic Technology Development Program(20001234).References
- Hripcsak G, Duke J D, Shah N, et al. Observational Health Data Sciences and Informatics (OHDSI): Opportunities for Observational Researchers. In Stud Health Technology Information. 2015;216:574-578. 2.
- Erickson B J, Korfiatis P, Akkus Z, et al. Machine Learning for Medical Imaging. Radiographics. 2017;32(2):505-515.
- Lai E C C, Ryan P, Zhang Y, et al. Applying a common data model to Asian databases for mutinational pharmacoepidemiologic studies: opportunities and challenges. Clinical Epidemiology. 2018;10:875.
- Park Y R, Shin S Y. Status and direction of healthcare data in Korea for artificial intelligence. Hanyang Medical Reviews, 2017;37(2):86-92
- Bidgood Jr W D, Horii S C, Prior F W, et al. Understanding and using DICOM, the data interchange standard for biomedical imaging. Journal of the American Medical Informatics Association. 1997;4(3):199-212.
Figures
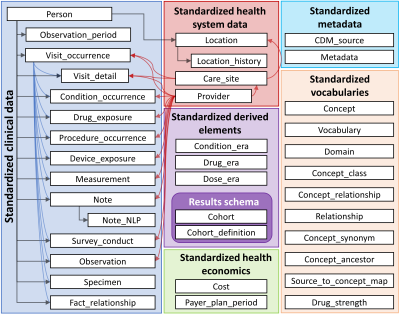
Figure 1. Schematic diagram of the relationship between tables
composing Observational Medical Outcomes Partnership (OMOP) – common data model
(CDM) (ver. 6.0). This figure provided a web-site “Observational Health Data
Sciences and Informatics – OHDSI,” available at https://ohdsi.org/.
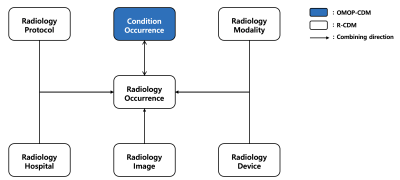
Figure 2. Schematic diagram of the relationship between tables
composing the radiology-common data model (R-CDM). Tables in white box (“Radiology_Occurrence,”
“Radiology_Image,” “Radiology_Protocol,” “Radiology_Modality,” “Radiology_Device,”
and “Radiology_Hospital”) Condition_Occurrence”) are those storing from DICOM
header. Table in blue box (“Condition_Occurrence”) is already existing in the
Observational Medical Outcomes Partnership-CDM and directly linked to the R-CDM
table “Radiology_Occurrence.”
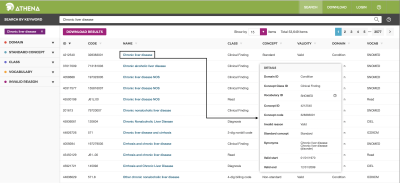
Figure 3. Athena web-site of “SNOMED” term for standardizing R-CDM terminology.
The web service is available at http://athena.ohdsi.org/search-terms/terms.
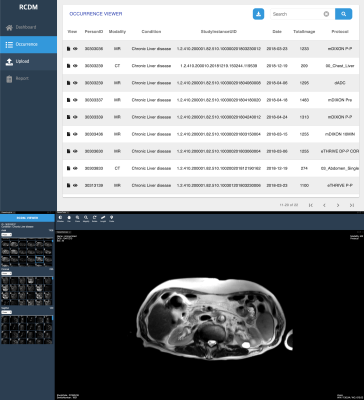
Figure 4. Occurrence
list viewer (upper) and DICOM image viewer (lower) on a web-based R-CDM
management system
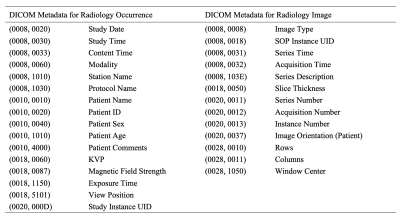
Table 1. The
contents of DICOM metadata for standardization of R-CDM