3651
An open-source graphical tool for interactive slice planning: Application to pseudo-continuous Arterial Spin Labeling
Jon-Fredrik Nielsen1, Luis Hernandez-Garcia1, and Douglas C. Noll1
1Biomedical Engineering, University of Michigan, Ann Arbor, MI, United States
1Biomedical Engineering, University of Michigan, Ann Arbor, MI, United States
Synopsis
Vendor-agnostic MR pulse sequence programming tools such as Pulseq or TOPPE allow rapid prototyping of complex MR sequences, but are not compatible with existing graphical slice planning interfaces on all supported vendor platforms. We introduce a simple open-source slice planning tool that allows any number of 3D rectangular regions-of-interest (ROIs) to be defined interactively and exported to a portable data file format (HDF5). We use this tool to prescribe both a 2D labeling plane in the neck and a 3D imaging volume in the brain for a vendor-agnostic implementation of 3D stack-of-spirals fast spin-echo (FSE) pseudo-continuous Arterial Spin Labeling.
Introduction
Implementing custom pulse sequences on commercial MRI systems generally requires low-level programming within a vendor’s proprietary software ecosystem, which can be technically challenging and time-consuming. However, once programmed, the custom sequence “behaves” much like a built-in sequence from the point of view of the operator; notably, the operator can prescribe the imaging volume (“slice planning”) in the same manner as for built-in sequences. On the other hand, commercial vendor consoles generally only permit a single 3D volume, or one stack of 2D slices, to be prescribed, which can be somewhat limiting for sequences such as pseudo-continuous Arterial Spin Labeling (pCASL), which requires prescription of both a 2D labeling plane and a 3D (or stack of 2D slices) imaging volume.Recently, alternative pulse programming environments have been proposed that are vendor-agnostic1-3, by allowing the user to define the sequence in Matlab (or Python) and export the sequence directly to a GE or Siemens scanner for execution via vendor-dependent, sequence-universal, “interpreter” executables. These environments promise to lower the barrier of entry into the pulse sequence development field for small to mid-sized laboratories with limited pulse programming expertise, and to foster collaborations across vendor platforms and sites. Unfortunately, programming sequences in this way does not generally take advantage of a vendor’s graphical slice planning capabilities, which we believe represents a significant barrier to adoption of these novel programming environments.
Here we introduce a simple, stand-alone, and open-source tool for graphical slice planning, that allows any number of 3D rectangular regions-of-interest (ROIs) to be defined interactively and exported to a portable data file format (HDF5). We also present a vendor-agnostic implementation of a 3D stack-of-spirals fast spin-echo (FSE) pCASL sequence, and demonstrate the use of our slice planning tool to prescribe both a 2D labeling plane in the neck and a 3D imaging volume in the brain for this sequence.
Methods
Interactive graphical slice planning: The Java source code for our graphical tool is freely available at https://github.com/toppeMRI/SlicePlanner. Each 3D ROI is represented internally as a rectangular box, and the intersection of this box with an arbitrary 2D viewing plane is calculated on-the-fly as the user interacts with the tool. The underlying geometric calculations are explicitly “hand-coded” and do not rely on Java libraries, and can be easily reused in other programming (e.g., C++) contexts. The graphical user interface (GUI) is written in JavaFX. Using the mouse, the user can interactively resize, move, and rotate each ROI, by manipulating the intersection of the ROI with axial, sagittal, and coronal views of the subject. In addition, the center slice within each ROI is displayed in separate windows, as an additional guide to ROI placement (a feature not commonly found on commercial scanners). The GUI also supports slice scrolling with the mouse wheel. Clicking the ‘Export ROIs’ button writes all ROIs to a single HDF5 file that can be loaded into Matlab with the ‘toppe.getroi()’ function in the TOPPE Matlab toolbox (https://toppemri.github.io) for subsequent use in the design of vendor-agnostic sequences based on, e.g., TOPPE1 or Pulseq2. This graphical tool can be run on commonly used personal computers, and does not rely on graphics processing units (GPU) or other advanced graphics features.Vendor-agnostic pCASL sequence: We implemented the stack-of-spirals FSE pCASL sequence in TOPPE. Sequence parameters: label duration 1.4s; adiabatic background suppression pulses; 3 spiral leafs per kz-platter; matrix size 80x80x20; FOV 24x24x6.1 cm3.
Results
We acquired a 3D localizer volume (also implemented in TOPPE) in a healthy volunteer on a GE MR750 3T scanner and loaded it into the GUI (Fig. 1). We then used the mouse to define the imaging and labeling planes, and used those ROIs to set the location and rotation of the labeling plane and imaging volume in the pCASL sequence. Figure 2 shows ASL perfusion maps obtained with the resulting vendor-agnostic pCASL sequence.Discussion and Conclusion
We have demonstrated that the proposed GUI allows interactive slice and label planning for a vendor-agnostic implementation of a 3D stack-of-spirals FSE pCASL sequence. Both the Java tool and the Matlab scripts for creating the pCASL sequence are fully open-source and freely available for download and further customization. Possible future applications of this work include body ASL and vessel-selective ASL, applications that require careful placement of the labeling plane as well as imaging volume.Acknowledgements
This work was supported by NIH Grants R21EB019653, R21AG061839, and in part, by NIH Grant R01 EB023618.References
- Nielsen JF, Noll DC. TOPPE: A framework for rapid prototyping of MR pulse sequences. Magn Reson Med. 2018; 79:3128-3134.
- Layton KJ, Kroboth S, Jia F, Littin S, Yu H, Leupold J, Nielsen JF, Stöcker T, Zaitsev M. Pulseq: A rapid and hardware-independent pulse sequence prototyping framework. Magn Reson Med 2017; 77:1544–1552.
- Ravi KS, Potdar S, Poojar P, Reddy AK, Kroboth S, Nielsen JF, Zaitsev M, Venkatesan R, Geethanath S. Pulseq-Graphical Programming Interface: Open source visual environment for prototyping pulse sequences and integrated magnetic resonance imaging algorithm development. Magn Reson Imaging 2018; 52:9-15.
Figures
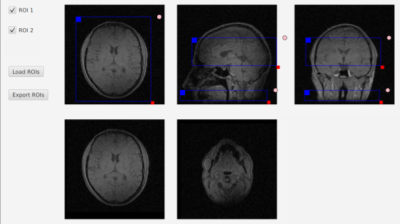
Figure 1. Graphical user interface (GUI) written in Java for defining (any number of) 3D rectangular ROIs. Here we prescribed a 3D imaging volume and a 2D spin labeling plane (top row) for a pcASL sequence implemented with a vendor-agnostic pulse programming software framework (TOPPE [1]). The underlying anatomical (localizer, or “scout”) image was also acquired with TOPPE. The bottom two frames show the center slice through each of the two ROIs.
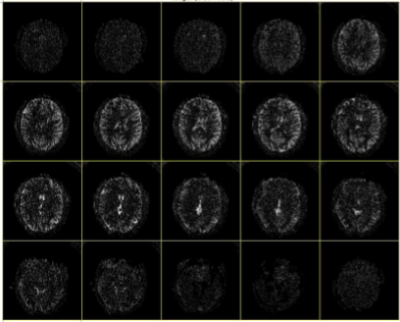
Figure 2. Pseudo-continuous ASL imaging results using a vendor-agnostic pulse sequence format (TOPPE) and the proposed graphical tool for positioning the labeling plane and imaging volume.