3649
Cross-vendor implementation of a Stack-of-spirals PRESTO BOLD fMRI sequence using TOPPE and Pulseq
Marina Manso Jimeno1,2, Sairam Geethanath1,2, Jon-Fredrik Nielsen3, and Douglas C. Noll3
1Columbia University, New York, NY, United States, 2Columbia Magnetic Resonance Research Center (CMRRC), New York, NY, United States, 3University of Michigan, Ann Arbor, MI, United States
1Columbia University, New York, NY, United States, 2Columbia Magnetic Resonance Research Center (CMRRC), New York, NY, United States, 3University of Michigan, Ann Arbor, MI, United States
Synopsis
TOPPE and Pulseq are two examples of open-source, vendor-agnostic frameworks for rapid prototyping and implementation of MR sequences. Both have demonstrated their ability to easily execute a variety of highly specific and customized sequences on different vendors. Cross-vendor translation of sequences generated by these two frameworks can enable multi-site and multi-vendor evaluation of a single sequence implementation. This study demonstrates this capability by porting a spiral-based fMRI sequence implemented in TOPPE on one vendor platform to Pulseq to run on another vendor. Results show comparable in vivo SNR and image contrast for Pulseq and TOPPE implementations.
Introduction
Magnetic Resonance (MR) Pulse Sequence Design (PSD) methods provide access to improvements in acquisition times, contrast and signal to noise ratio.1 For this reason, the flexibility and pace of the process from prototyping to implementation is key.2 Open source, vendor-neutral tools such as TOPPE3 and Pulseq4 deliver these requirements. They allow sequence design and deployment on multiple vendor platforms.The two frameworks can work in a complementary way in specific scenarios.4 For example, TOPPE is the Pulseq’s interpreter for GE (GE Healthcare, WI). They construct the sequences by putting together basic building blocks that are usually repeated over time. Ultimately, the code output condenses these blocks into a vendor-independent file that will run in the system through an interpreter module. Figure 1 summarizes this workflow.
Recent studies have shown the feasibility of these methods by demonstrating different sequences on different vendor platforms.3,4 However, the cross-vendor implementation of a particular sequence for verified identical execution has not been performed. We address this in our work.
We utilize a stack-of-spirals PRESTO5 sequence for resting-state functional MRI (“spiral fMRI”). It allows dynamic T2* weighed contrast with high temporal resolution and short total acquisition time.5
Methods
We used the results of the original TOPPE implementation of the spiral fMRI sequence on a GE MR750 3T for benchmarking. First we “translated” the MATLAB (The MathWorks, Inc.) -based TOPPE code to the Python version of Pulseq (pyPulseq6). We directly imported gradient and RF waveforms from TOPPE to avoid design errors and achieve the most similar execution. These waveforms were scaled and interpolated to account for differences in units, raster times and safety limits between vendors. Table 1 lists these cross-vendor hardware constraints and units. The sequence acquisition parameters were: 10° flip angle, 16.7 ms TR, 32 ms TE (Note that PRESTO technique is characterized by a TE larger than TR), Field of View (FOV) of 240 x 240 x 180 mm3 with an isotropic resolution and a reconstruction matrix size of 72x72x54.The spiral fMRI acquisition had three modules: fat-saturation, RF slab excitation (which included the PRESTO gradients) and read-out. Figure 3 shows the pulse sequence diagram. The acquisition consisted of single spiral-in shots that rotate at every platter and frame, each shot with 966-time points. 54 platters built a volume at one time frame and had an acquisition time of 0.9 seconds (Figure 4A). Combining three frames, three leaves with different angles come together at each platter, creating a fully-sampled k-space volume (Figure 4B) in 2.7 seconds.
We tested the sequence implementation both in vitro and in vivo. We used the FBIRN (Functional Biomedical Informatics Research Network) phantom, which helps with the evaluation of the sequence’s Signal to Noise-Ratio (SNR). In this case, we computed SNR as the ratio of the mean intensity over the phantom region and the standard deviation of the background. We segmented two mutually exclusive Regions of Interest (ROIs) for the calculation: 1. a circle that covers the phantom area and 2. the remaining region of the image. Finally, we acquired the images of a healthy volunteer’s brain as part of an approved IRB study. The experiments were performed on a Siemens Prisma Fit 3T (Siemens Healthineers, Erlangen; Vendor 2). Image reconstruction was performed using the Non-Uniform Fourier Transform (NUFFT) code available in the Michigan Image Reconstruction Toolbox (MIRT).7 The reconstruction code for this data acquisition is also available online.8
Results and discussion
We demonstrated cross-vendor implementation after the sequence successfully ran on vendor 2. The acquisition time for a fully sampled volume (2.7 seconds) concurs with the value stated for vendor 1, which was 2.9 s. This difference can be attributed to the change in TR, being 18 ms on vendor 1 and 16.7 ms on vendor 2. The SNR calculated from the in vitro experiment was 57 dB.In vivo results (Figure 5B) show that the contrast (T2*) and general appearance of the images are comparable to the results previously generated from vendor 1 and reported in Fig. 4 of ref3. Off-resonance artefacts are visible in the prefrontal cortex area of the brain (Figure 5B, red arrow).
The code to generate the sequence file(s) that execute on the MRI machines for both frameworks is available in a GitHub repository.8
Conclusion
We have demonstrated the feasibility of cross-vendor implementation of the spiral PRESTO sequence. Our in vitro studies show an acceptable SNR while in vivo images present the same contrast as the originally reported results. We will incorporate k-space undersampling to the sequence. After that, we will be able to assess the sequence’s cross-vendor reproducibility. Current and future work also involves incorporating off-resonance correction and integrating with standard resting-state fMRI processing pipelines.Acknowledgements
- Fast Functional MRI with sparse sampling and model-based reconstruction, National Institute of Biomedical Imaging and Bioengineering, 1R01EB023618-01, PI: Noll, Fessler; UM SUBK00009808, PI: Geethanath;
- Zuckerman Institute Technical Development Grant for MR, Zuckerman Mind Brain Behavior Institute, Grant Number: CU-ZI-MR-T-0002; PI: Geethanath;
References
- Magland, J. F., Li, C., Langham, M. C., & Wehrli, F. W. (2016). Pulse sequence programming in a dynamic visual environment: SequenceTree. Magnetic resonance in medicine, 75(1), 257–265. doi:10.1002/mrm.25640
- Ravi, K. S., Potdar, S., Poojar, P., Reddy, A. K., Kroboth, S., Nielsen, J. F., Zaitsev, M., Venkatesan, R., Geethanath, S. (2018). Pulseq-Graphical Programming Interface: Open source visual environment for prototyping pulse sequences and integrated magnetic resonance imaging algorithm development. Magnetic Resonance Imaging, 52, 9-15. doi:10.1016/j.mri.2018.03.008.
- Nielsen, J. F., & Noll, D. C. (2018). TOPPE: A framework for rapid prototyping of MR pulse sequences. Magnetic resonance in medicine, 79(6), 3128–3134. doi:10.1002/mrm.26990
- Layton, K. J., Kroboth, S., Jia, F., Littin, S., Yu, H., Leupold, J., Nielsen, J., Stöcker, T. and Zaitsev, M. (2017), Pulseq: A rapid and hardware‐independent pulse sequence prototyping framework. Magn. Reson. Med., 77: 1544-1552. doi:10.1002/mrm.26235
- van Gelderen, P., Duyn, J. H., Ramsey, N. F., Liu, G., & Moonen, C. T. (2012). The PRESTO technique for fMRI. NeuroImage, 62(2), 676–681. doi:10.1016/j.neuroimage.2012.01.017
- Ravi et al., (2019). PyPulseq: A Python Package for MRI Pulse Sequence Design. Journal of Open Source Software, 4(42), 1725, https://doi.org/10.21105/joss.01725
- https://web.eecs.umich.edu/~fessler/code/mri.htm
- https://github.com/imr-framework/spiral-fmri
Figures
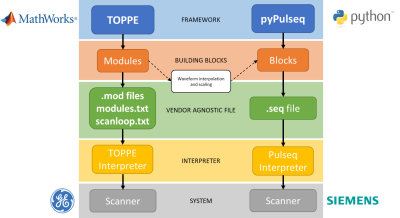
Figure 1: Comparison of TOPPE’s and Pypulseq’s pipelines from sequence coding to scanner implementation. The architecture of both frameworks is similar having both the same main components. The translation from one another only requires an extra step for waveform interpolation and scaling.
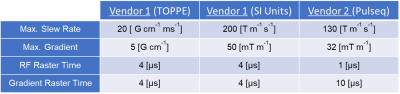
Table 1: System gradient/slew rate limits and RF/gradient raster time specifications for both vendors. Parameters for vendor 1 are converted to SI units for a better comparison.
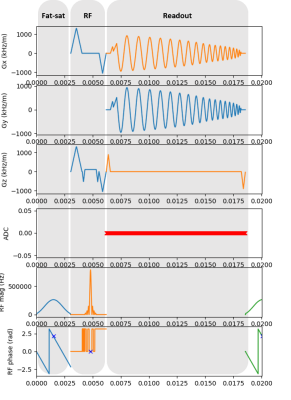
Figure 3: pyPulseq’s Pulse sequence diagram. From top to bottom: gradients along the x-axis (Gx), y-axis (Gy), and z-axis (Gz) in kHz per meter, ADC, RF phase and RF magnitude in Hz and radians, respectively. The three grey areas represent the equivalent TOPPE modules for fat saturation pulse, RF slab excitation and spiral readout gradients.
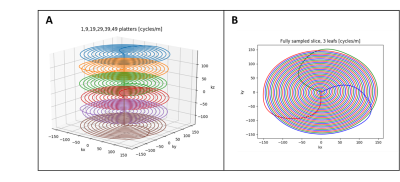
Figure 4: K-space trajectory plots. A) 3D visualization of platters (6 out of 54) that create a time frame. The spiral-in leaf rotates at every platter. B) In-plane plot of the trajectory after combining three frames to construct a fully sampled volume.
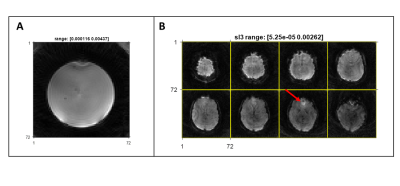
Figure 5: A) Slice of a fully sampled volume of the FBIRN phantom acquired using the spiral fMRI sequence in vendor 2. B) Eight slices of a fully sampled volume of the brain of a healthy volunteer acquired using the spiral fMRI sequence in vendor 2. The red arrow points to artifacts due to off-resonance effect on the prefrontal cortex.