3647
Python pulse sequence development kit for a fast MRI simulator1MRIsimulations,Inc., Tokyo, Japan
Synopsis
A software tool that can efficiently describe MRI pulse sequences in Python has been developed for a fast MRI simulator. The essential part of this tool consists of application program interface (API) developed in C++, and any pulse sequence can be written by calling the API from a Python program. Using this tool, several pulse sequences including magnetic resonance fingerprinting were developed, and their usefulness was evaluated by performing MRI simulations for numerical phantoms. As a result, the tool developed in this study was shown to be very efficient in developing MRI pulse sequences.
Introduction
MRI vendors provide their own pulse sequence development environments using high level languages such as C and C ++, and these environments are widely used for pulse sequence developments in the MRI community. On the other hand, a pulse sequence development framework independent of MRI vender hardware has been proposed1. In 2017, we published a fast MRI simulator2 that operates exactly the same pulse sequence as some MRI systems. The pulse sequence format2-4 supported by the simulator had an advantage that it was very easy to develop pulse sequences, but there was a problem that it was very complicated to develop pulse sequences with many arbitrary RF and gradient waveforms. To solve this problem, we developed a software tool (Python pulse sequence development kit: PSDK) that can efficiently describe pulse sequences in Python. The usefulness of this tool was evaluated by MRI simulations using various pulse sequences.Python pulse sequence development kit
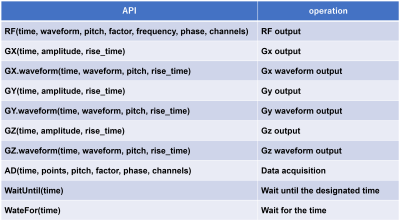
Figure 1 shows the application program interface (API) that is the interface between Python and the MRI simulator developed in C++. It consist of “RF” for controlling RF pulses, “GX”, “GY”, and “GZ” for controlling gradient amplitudes with linear rise or fall, “GX.waveform”, “GY.waveform”, and “GZ.waveform” for controlling arbitrary gradient waveforms, “AD” for data acquisition, “WaitFor” and “WaitUntil” for sequence timing control. Since both scalar values and tuples can be used for the arguments of the API functions, flexible description of control structures is possible. Since all of the Python statements are used for the sequence description, various pulse sequences can be written with the small number of the API functions. To demonstrate the usefulness of the PSDK, MRI simulations were performed using numerical phantoms and the fast MRI simulator4 running on a graphical processing unit (GeForce GTX-1070 for laptop, NVIDIA, Santa Clara, CA).Results
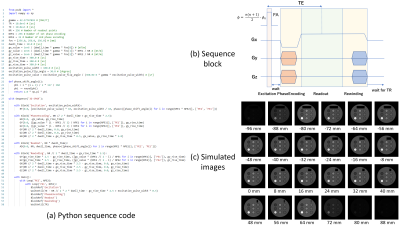
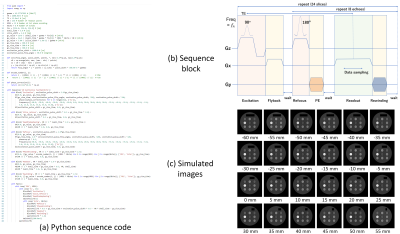
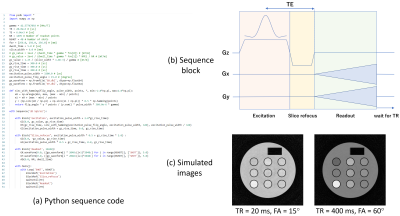
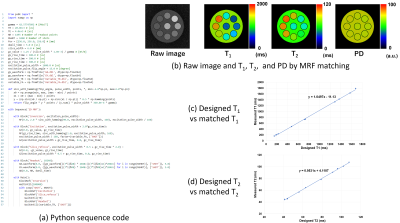
Figures 2-4 shows (a) pulse sequence codes written in Python, (b) pulse sequence blocks, and (c) simulation results for a 3D RF spoiled gradient-echo (GRE), a multi-slice fast spin-echo (FSE), and a 2D spiral imaging sequences. The Python code for the 3D RF spoiled GRE sequence consists of ‘Excitation’, ‘PhaseEncoding’, ‘Readout’, and ‘Rewinding’ blocks and a dual sequence loop with loop variables of ‘PE2’ and ‘PE1’ as described by Main(). The simulated result clearly shows 3D T1 weighted images. The Python code for the multi-slice FSE sequence consists of ‘Excitation’, ‘Slice_refocus’, ‘ReadPredephasing’, ‘Refocus’, ‘PhaseEncoding’, ‘Readout’, and ‘Rewinding’ blocks and a triple loop with loop variables of ‘PE1’ (Phase encoding), ‘SL’ (multiple slice), and ‘Echo’ (multiple echo) as described by Main(). The simulated result clearly shows multiple-slice T2 weighted images. The Python code for the spiral sequence consists of ‘Excitation’, ‘Slice_refocus’, and ‘Readout’ blocks and a single loop with a loop variable of ‘SHOT’ as described by Main(). In the ‘Readout’ block, the data acquisition was performed under the application of Gx and Gy field gradients described by the GX.waveform and GY.waveform API functions. The waveforms of the Gx and Gy gradients were those of the 48-shot variable density spiral trajectory designed by the formalism developed by Kim et al5. The waveforms were calculated beforehand and input from the “Gx.dbl” and “Gy.dbl” files. The simulated result clearly shows the spiral images as expected from the sequence parameters. Figure 5 shows (a) pulse sequence codes written in Python, (b) raw image data and T1, T2, and proton density (PD) images obtained from a dictionary matching, (c) matched T1 plotted against the designed T1, and (d) matched T2 plotted against the designed T2 for the magnetic resonance fingerprinting (MRF) sequence. The Python code for the MRF sequence consists of ‘Inversion’, ‘Excitation’, ‘Slice_refocus’, and ‘Readout’ blocks and a single loop with a loop variable of ‘SHOT’ as described by Main(). The variable TR and variable flip angle (FA) were calculated beforehand and input from “variable_TR.dbl” and “variable_FA.dbl” files. TR was changed between 15 and 20 ms and randomized using perlin noise. FA was sinusoidally changed between 0-200, 250-450, 500-700, and 750-950 shots of the 1,000 times spiral acquisition. In the MRF acquisition, the 48-shot spiral trajectories as used in Fig.4 were used and signal sampling was performed while increasing the trajectory rotation angle at intervals of 7 trajectories (52.5°) at each TR. Image reconstruction was performed for the seven consecutive datasets using the sliding window reconstruction technique. The PD, T1, and T2 images obtained from the dictionary matching demonstrate the correctness of the MRF sequence.Discussion
By using the API developed in this study, we succeeded in describing the pulse sequence of 3D spoiled GRE, 2D multi-slice FSE, 2D spiral, and 2D MRF with as few as 50-90 Python statements. The largest advantage of our pulse sequence development tool is that the MRI pulse sequence can be written in the Python statements themselves, which is very different from the development tools published so far. This tool was developed for our MRI simulator, but if API is developed for a real MRI machine, the same pulse sequence can be used simultaneously for the real machine and the simulator. In conclusion, we succeeded in development of a software tool efficiently describing pulse sequences for a fast MRI simulator in Python statements and the API.Acknowledgements
No acknowledgement found.References
[1] Layton KJ, Kroboth S, Jia F, Littin S, Yu H, Leupold J, Nielsen JF, Stöcker T, Zaitsev M. Pulseq: a rapid and hardware-independent pulse sequence prototyping framework. Magn Reson Med 2017;77:1544–1552.
[2] Kose R, Kose K. BlochSolver: A GPU-optimized fast 3D MRI simulator for experimentally compatible pulse sequences. J Magn Reson 2017;281:51-65.
[3] Hashimoto S, Kose K, Haishi T. Development of a pulse programmer for magnetic resonance imaging using a personal computer and a high-speed digital input–output board, Rev. Sci. Instrum 2012;83:053702.
[4] Kose R, Setoi A, Kose K. A fast GPU-optimized 3D MRI simulator for arbitrary k-space sampling. Magn Reson Med Sci, 2019;18:208-218.
[5] Kim D, Adalsteinsson, Spielman DM. Simple analytic variable density spiral design. Magn Reson Med 2003;50:214–219.
Figures