3645
Reconstream: A Software Framework for Developing Image Reconstruction Algorithms in Scientific Research Activities1Advanced Technology Research Department, Research and Development Center, Canon Medical Systems Corporation, Kanagawa, Japan
Synopsis
Existing software tools for assisting scientific research mainly focus on increasing reproducibility of primitives and/or integrated algorithms (exchangeable reproducibility). While exchangeable reproducibility is clearly important, reproducing the outputs using past configurations again is also important in daily research activities (local reproducibility).
The author proposes a new software framework named reconstream. To improve local reproducibility, reconstream implemented a revision manager for reproducing past configurations easily. To improve exchangeable reproducibility, the author reviewed advantages of individual software tools and implemented them in the reconstream framework.
Experimental results demonstrate several examples using various features of reconstream.
INTRODUCTION
For acquisition and reconstruction of MR images, many studies use algorithms with high complexity. It is not easy to reproduce and improve state-of-the-art algorithms since they often consist of many primitives and hyperparameters. To increase reproducibility of primitives and/or integrated algorithms, several research teams developed open-source software tools for reading/writing k-space data1, providing primitives for building complicated algorithms easily2, reconstructing k-space data as live streams3, and editing a set of primitives (a flow) of reconstruction interactively4. These tools mainly focused on providing ways for reproducing existing algorithms. To distinguish from a term mentioned later, this paper calls this reproducibility as exchangeable reproducibility.While exchangeable reproducibility is clearly important, in daily research activities it is also important to reproduce the outputs using past revisions of data flows. In the software engineering field, there are several implementations of version control systems (VCS) such as cvs, subversion, git and mercurial for reproducing past revisions of software. The author named the capability of such reproduction as local reproducibility. For example, it is useful to compare performance using a new flow to performance using past flows. However, researchers rarely report the version of the open-source software they are working with for reproducibility.
In addition, there seems to be no tools which support both streaming reconstruction and graphical edit tools simultaneously. Both of functionalities are useful for developing new algorithms. However, existing tools support at most one of these functionalities.
To overcome limitations of existing open-source software tools, the author proposes a new software framework named reconstream. The primary motivation of developing reconstream is to implement both exchangeable and local reproducibility for improving research activities of MRI reconstruction. In addition, the motivation of developing reconstream includes to provide a tool which supports both streaming and interactive reconstruction simultaneously.
METHODS
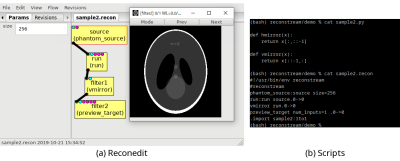
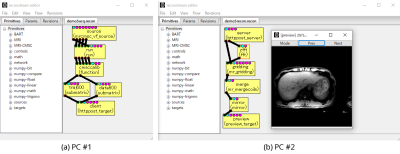
Reconstream is a multiplatform framework for editing and running a flow and is written mainly in Python5. A flow consists of inputs, outputs, a set of primitives, connections between primitives, hyperparameters, and an entry point for running the flow. Reconstream implemented three tools: reconedit which is an interactive flow editor using wxPython6, a non-interactive command-line program for running flows, and a library for managing flows.To improve local reproducibility in daily research activities, reconedit automatically keeps past flows as revisions. The flows that were run successfully can be reused later. In addition, flows can be saved as a script with parameters which can be set in both a non-interactive way as command-line arguments and an interactive way as a dialog of reconedit. A script can be generated by reconstream as a file with a shebang and executable file permissions. A script can also be created or edited using text editors.
To improve exchangeable reproducibility, the author reviewed advantages of individual software tools and implemented them in reconstream as shown in Table 1. For reproducing existing MRI algorithms, reconstream implemented primitives for running NumPy functions7, a fast Fourier transform (FFT) with a swap operation known as fftshift, a gridding algorithm8, a non-uniform FFT9, primitives for parallel imaging (PI)10,11, a primitive for estimating missing cardiac triggers, a primitive for correcting errors of radial trajectories, and some other algorithms. Reconstream also implemented primitives for calling commands of BART2 seamlessly. To implement a new primitive algorithm easily, reconstream has the capability to import functions which are developed independently from reconstream if their arguments and return values are supported types.
For reading k-space data, reconstream implemented primitives using file readers in BART and a primitive using a vendor-specific parser for Canon Medical Systems MRI. In order to run primitives on GPU, primitives for calling NumPy functions transparently call CuPy functions12 in the cases that inputs are instances of CuPy classes.
RESULTS
Figures 1-4 demonstrate that reconstream can be used for both prototyping scientific algorithms visually, and running developed algorithms on various platforms including systems with no human machine interfaces. Figure 1 shows a screenshot of reconedit. Run primitives in a flow are called sequentially when the flow is run. In another screenshot shown in Figure 2, a past flow is reused on reconedit. In Figure 3, reconstream is used for running a user-defined primitive. Figure 4 demonstrates an image reconstruction algorithm as a live stream over HTTP. A machine converts vendor-specific data to vendor-independent raw data and sends it. Another machine receives the converted raw data and reconstructs it.DISCUSSION
It is well-known that reproduction of software revisions is important for daily development in the software development field. While the concept of local reproducibility is new in research tools of MRI, the concept is already used in research activities by researchers’ own ways of working.Reconstream is a general framework for prototyping arbitrary streaming applications. While the current version of reconstream is mainly focused on reconstruction of MR data, reconstream can be used for other applications. Development of other applications such as prototyping pulse sequences13 remains as future work.
CONCLUSION
The author proposes a new software framework named reconstream. To improve local reproducibility, reconstream implemented a revision manager for reproducing past configurations easily. To improve exchangeable reproducibility, the author reviewed advantages of individual software tools and implemented them to reconstream.Acknowledgements
No acknowledgement found.References
1. Inati SJ, Naegele JD, Zwart NR, et al. ISMRM Raw data format: A proposed standard for MRI raw datasets. Magn Reson Med 2016; 77(1): 411–421.
2. Uecker M, Ong F, Tamir J, et al. Berkeley advanced reconstruction toolbox. In Proc. Intl. Soc. Magn Reson Med 23: 2486.
3. Hansen MS, Sørensen TS. Gadgetron: An open source framework for medical image reconstruction. Magn Reson Med 2013; 69(6): 1768-1776.
4. Zwart NR, Pipe JG. Graphical programming Interface: A development environment for MRI methods. Magn Reson Med 2015; 74(5): 1449–1460.
5. https://www.python.org/
6. https://wxpython.org/
7. https://www.numpy.org/
8. Jackson JI, Meyer CH, Nishimura DG, Macovski A. Selection of a convolution function for Fourier inversion using gridding. IEEE Trans Med Imaging. 1991;10(3):473-478.
9. Keiner J, Kunis S, Potts D. Using NFFT 3 - a software library for various nonequispaced fast Fourier transforms. ACM Trans Math Software. 2009; 36 (19): 1-30.
10. Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: Sensitivity encoding for fast MRI. Magn Reson Med 1999; 42: 952–962.
11. Griswold MA, Jakob PM, Heidemann RM, Nittka M, Jellus V, Wang J, Kiefer B, Haase A. Generalized autocalibrating partially parallel acquisitions (GRAPPA). Magn Reson Med 2002; 47: 1202–1210.
12. https://cupy.chainer.org/
13. Ravi KS, Potdar S, Poojar P, et al. Pulseq-graphical programming interface: Open source visual environment for prototyping pulse sequences and integrated magnetic resonance imaging algorithm development. Magn Reson Med 2018; 52: 9-15.
Figures