3644
Graphical sequence visualization and development in the dynamic platform-independent framework gammaSTAR (γ*): A first prototype
Simon Konstandin1, Cristoffer Cordes1, and Matthias Günther1,2
1MR Physics, Fraunhofer MEVIS, Bremen, Germany, 2MR-Imaging and Spectroscopy, Faculty 01 (Physics/Electrical Engineering), University of Bremen, Bremen, Germany
1MR Physics, Fraunhofer MEVIS, Bremen, Germany, 2MR-Imaging and Spectroscopy, Faculty 01 (Physics/Electrical Engineering), University of Bremen, Bremen, Germany
Synopsis
MR sequence development is usually carried out by means of manufacturer-specific frameworks that do not allow an easy sequence transfer to scanners from other manufacturers. Sequence development in the recently presented framework is performed by scripting that can be time consuming and confusing for sophisticated MR sequences. Here, a module editor prototype is presented that provides a graphical sequence representation and should allow for a fast and relatively easy-to-understand MR sequence development in the future. Functionality could already be demonstrated by exchanging the rf pulse and implementing a parallel acquisition technique into a gradient echo sequence in a few steps.
Introduction
MR sequence development is usually carried out by means of manufacturer-specific frameworks that do not allow an easy sequence transfer to scanners from other manufacturers. Thus, multicenter studies mostly use only scanners of one type and research sequences cannot be easily distributed and further developed. Various software tools have been introduced to overcome this problem.1-5 Existing disadvantages such as additional required compilation steps and the unavailability of changing sequence parameters during the scan have been solved both by a recently developed framework gammaSTAR (γ*).6-8 So far, sequence development in this framework was performed by scripting that can be time consuming and confusing for sophisticated MR sequences.In this abstract, a prototype of a graphical module editor is presented, which provides a clear sequence visualization as well as fast editing/ inserting/ removing of existing modules for a graphical MR sequence development.
Methods
The module editor was implemented into the framework gammaSTAR (γ*), which already provides a set of standard 2D/3D sequences and modules.7 The presented module editor was developed using the D3.js framework for visualization and interaction, embedded into the Quasar web frontend. The state of the editor is synchronized with the other sequence development tools, such that any change propagates throughout all views.Results & Discussion
The functioning of the module editor is demonstrated by exchanging a rf pulse with subsequent implementation of a reference scan for GRAPPA9. In Figure 1, the design of the presented module editor is shown for the FLASH10 sequence. Sequence elements (e.g. loops, rf/gradient events) are represented by green nodes. There are several options to visualize the sequence tree more clearly (e.g., collapse/expand subtrees, show certain levels only, show subtrees) and to change the sequence structure. Modules/subtrees can be deleted, inserted anywhere in the tree and replaced by other modules, if they are of the same type. To transform the sinc rf pulse into a multiband pulse, the sinc pulse must be replaced first, whereas the orange color indicates the rf type (Figure 2). By means of the module search (see ‘RF SINC SMS’ in Figure 2), a new module can be created with the ‘new mod’ button. The rf pulse can easily be inserted via drag and drop, whereas the corresponding gradient is automatically updated. After refreshing the sequence and changing the rf protocol parameters, the multiband pulse can be seen in the sequence plot (Figure 3 right). A similar approach is performed to insert the GRAPPA reference scan (Figure 3), which leads to the final result shown in Figure 4 after refreshing the sequence. The acquisition scheme itself does not have to be adapted for parallel acquisition since a different module already determines the trajectory information among others from GRAPPA protocol parameters is inherently available in every sequence. The automatic generation and linking of new modules’ parameters (e.g. multiband factor, GRAPPA factor) are currently under development and were done manually for this abstract.Conclusion
A module editor prototype was presented that provides a graphical sequence representation and should allow for a fast and relatively easy-to-understand MR sequence development in the future. Functionality could already be demonstrated by exchanging the rf pulse and implementing a parallel acquisition technique into a gradient echo sequence in a few steps.Acknowledgements
All funding for this study was provided by the internal Attract (600172) funding program of the German Fraunhofer-Gesellschaft.References
- Jochimsen TH & von Mengershausen M. ODIN-Object-oriented development interface for NMR. J Magn Reson 2004;170(1):67-78.
- Stöcker T, Vahedipour K, Pflugfelder D, Shah NJ. High-performance computing MRI simulations. Magn Reson Med 2010;64(1):186-93.
- Magland JF, Li C, Langham MC, Wehrli FW. Pulse sequence programming in a dynamic visual environment: SequenceTree. Magn Reson Med 2016;75(1):257-65.
- Layton KJ, Kroboth S, Jia F, Littin S, Yu H, Leupold J, Nielsen JF, Stöcker T, Zaitsev M. Pulseq: A rapid and hardware-independent pulse sequence prototyping framework. Magn Reson Med 2017;77(4):1544-52.
- Nielsen JF & Noll DC. TOPPE: A framework for rapid prototyping of MR pulse sequences. Magn Reson Med 2018;79(6):3128-34.
- Cordes C, Konstandin S, Porter D, Günther M. Portable and platform-independent MR pulse sequence programs. Magn Reson Med doi:10.1002/mrm.28020.
- https://gamma-star.mevis.fraunhofer.de.
- Konstandin S, Cordes C, Günther M. Dynamic platform-independent MRI vs. manufacturer’s implementations. Proc Intl Soc Mag Reson Med 27 p.4841.
- Griswold MA, Jakob PM, Heidemann RM, Nittka M, Jellus V, Wang J, Kiefer B, Haase A. Generalized autocalibrating partially parallel acquisitions (GRAPPA). Magn Reson Med 2002;47(6):1202-10.
- Haase A, Frahm J, Matthaei D, Hanicke W, Merboldt KD. FLASH imaging. Rapid NMR imaging using low flip-angle pulses. J Magn Reson (1969) 1986;67(2):258-66.
Figures
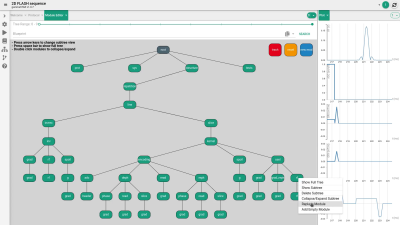
FIG. 1. Design of the module editor and options to change the
sequence structure. The green nodes correspond to the sequence elements like
loops and rf/gradient events (see text for detailed description).
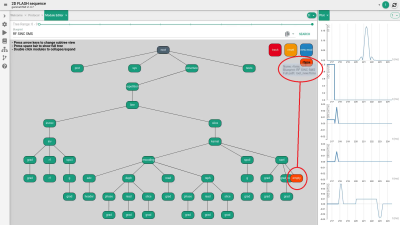
FIG. 2. Conversion of the sinc rf pulse into a multiband pulse
by choosing and creating the new module from a search list and using it via
drag and drop (orange ellipses, see text for detailed description).
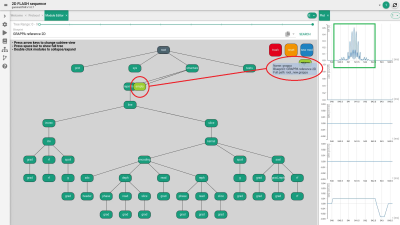
FIG. 3. The multiband rf pulse can be seen in the sequence plot (green
square) after sequence refresh, whereas the corresponding gradient is automatically updated. A
GRAPPA reference scan for parallel acquisition can be implemented in a similar
way as it was done for the rf pulse (orange ellipses, see text for detailed
description).
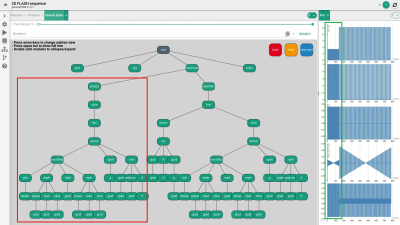
FIG. 4. Final sequence structure with easily and fast
integrated GRAPPA module (orange and green squares, see text for detailed
description).