3641
Are deep learning MR reconstruction models robust against adversarial attacks?
Taohui Xiao1, Cheng Li1, Haoyun Liang1, Hairong Zheng1, and Shanshan Wang1
1Paul C. Lauterbur Research Center for Biomedical Imaging, SIAT, Chinese Academy of Sciences, Shenzhen, P.R.China, Shenzhen, China
1Paul C. Lauterbur Research Center for Biomedical Imaging, SIAT, Chinese Academy of Sciences, Shenzhen, P.R.China, Shenzhen, China
Synopsis
This paper investigates the robustness of deep learning MR reconstruction models for adversarial attacks like new lesions, different anatomy and noise pollutions. Specifically, three popular MR reconstruction algorithms were selected to investigate this issue. Experimental results show that model-based deep learning MR reconstruction method is relatively more robust than end-to-end data-driven reconstruction networks when transfer to other organs or face new lesions. Data-driven approaches can achieve better results when the testing images follow similar distributions as the training images. Severe noise can be a big issue for both deep learning methods and the traditional method.
Introduction
Deep learning has achieved great success lately. However, adversarial attacks have been identified to be able to affect the security of deep learning algorithms1. Some small changes occur in the dataset that are barely visible to human eyes can lead to very different outputs of the deep learning algorithms. Meanwhile, there are many studies applying deep learning to the field of medical image reconstruction2,3. Although it has not been investigated, adversarial attacks certainly exist in the field of reconstruction. Current research on deep learning-based MR image reconstruction focuses on training and testing with data from the same scan regions, such as the brain, knee, or heart. And the training data are collected mainly from healthy volunteers. We call them control samples. However, in the actual clinical situations, the majority of the scan objects are patients with lesion or noisy structures, which have never been observed for networks trained with control samples. Therefore, it is extremely important to know if reconstruction models can handle this issue. In this work, we design a series of experiments to verify the robustness of the reconstruction model, including transferring the models to reconstruct MR images of other organ tissues and testing the impact of the adversarial sample with new lesion structures and noise.Method
We choose two advanced deep learning algorithms to investigate. One is the model based deep learning method: VN 3, and the other is the end-to-end network: Unet4. To compare the reconstruction performance of the methods, the classical SPIRiT algorithm is used as a baseline. Our study can be divided into four sections. The first step is to verify and ensure the reconstruction performance of the reconstruction models. We train and test VN and Unet on the knee dataset or brain dataset as shown in config1 in figure 1. The second step is to test the transfer performance of the models, as shown in config2 in figure 1. In the third step, adversarial attack experiments are carried out by adding adversarial samples with lesions. MR images of the femur and brain containing lesions are tested. The last section is to validate the robustness of the models by testing adversarial samples with noise. We manually add Gaussian white noise to the image and adjust the amount of noise by changing the value of standard deviations. The overall framework of our study is shown in figure 1.Experiment
The data used in the experiments include 15-channel knee data provided by VN (https://github.com/VLOGroup/mri-variationalnetwork), our inhouse 12-channel brain data, the ATLAS data (http://fcon_1000.projects.nitrc.org/indi/retro/atlas_download.html), and the femur images (https://radiopaedia.org/cases/breast-cancer-metastasis-femur-mri?lang=us). The 15-channel data were tested using channel compression code in ESPIRiT to compress it to 12 channels. We used ESPIRiT to estimate the sensitivity map of 12-channel brain data and used it as the sensitivity map of ATLAS data to simulate the 12-channel data. Similarly, the sensitivity map of knee data was used to simulate the sensitivity map of femur image. In addition, combined with the segmentation label provided by ATLAS, we extracted the lesion tissues in the corresponding images and added them to the brain data we collected to simulate the lesion tissues. Studies have shown that the difference in test results is rarely related to contrast 5. Therefore, in this work we ignore the difference between different contrasts. The knee data tested is PD-weighted, the brain data is T2-weighted data, ATLAS is T1-weighted, and the femur is T2-weighted. The undersampling pattern used in all the experiments is 1Drandom sampling (33%).Results and Discussion
Figure 2 shows the experimental results of config1 and config2 in Figure 1. The results of SPIRiT are used as a baseline. It can be seen that VN and Unet can reconstruct the same structure very well when training and testing on the data from the same regions. When training and testing with images from different structures, the performances of both methods are decreased. But VN is more robust than Unet in this case. Figure 3 shows the results of config3 in Figure 1. The test data are MR images with lesions. According to Figure 3, Unet presents the worst reconstruction performance in all images. VN and SPIRiT have their own advantages. This further proves that the VN model is relatively more robust than Unet and SPIRiT is relatively more stable. Figure 4 and Figure 5 are results of config4 in Figure 1. The results show that the three algorithms have very poor reconstruction performances with increased noise. At the same noise level, VN and Unet seems to be able to suppress more noise.Conclusion
Both model-based deep learning MR image reconstruction and end-to-end network were capable of reconstructing testing images which have the same data distribution as the training dataset with high image qualities. However, when transferring to other organs and tissues, the generalization abililty of the classical method SPIRiT is more stable compared to deep learning methods. Furthermore, model-based deep learning reconstruction method was relatively more robust than the data-driven end-to-end deep learning network. Data-driven approaches could achieve better results when the testing images follow similar distributions as the training images. Severe noise can be a big issue for both deep learning methods and the traditional method.Acknowledgements
This research was partly supported by the National Natural Science Foundation of China (61601450, 61871371, 81830056), Science and Technology Planning Project of Guangdong Province (2017B020227012, 2018B010109009), the Basic Research Program of Shenzhen (JCYJ20180507182400762), Youth Innovation Promotion Association Program of Chinese Academy of Sciences (2019351).References
[1] Madry A, Makelov A, Schmidt L, et al. Towards deep learning models resistant to adversarial attacks [J]. arXiv preprint arXiv:1706.06083, 2017. [2] Wang S, Su Z, Ying L, et al. Accelerating magnetic resonance imaging via deep learning[C]//2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI). IEEE, 2016: 514-517. [3] Hammernik K, Klatzer T, Kobler E, et al. Learning a variational network for reconstruction of accelerated MRI data[J]. Magnetic resonance in medicine, 2018, 79(6): 3055-3071. [4] Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation[C]//International Conference on Medical image computing and computer-assisted intervention. Springer, Cham, 2015: 234-241. [5] Knoll F, Hammernik K, Kobler E, et al. Assessment of the generalization of learned image reconstruction and the potential for transfer learning[J]. Magnetic resonance in medicine, 2019, 81(1): 116-128.Figures
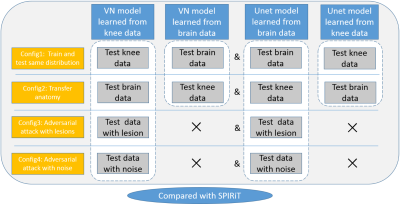
Fig.
1 Experimental scheme diagram
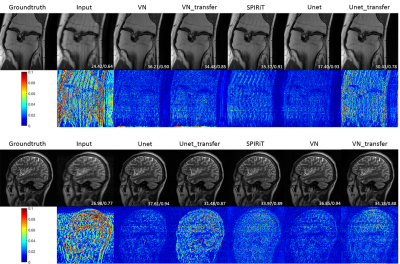
Fig. 2 Comparison of VN, Unet, SPIRiT
reconstruction results. VN_transfer and Unet_transfer represent the transfer
test.
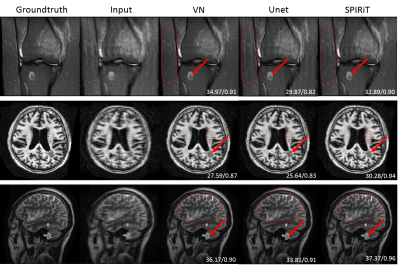
Fig. 3 Reconstruction of MR lesion images
by VN, Unet and SPIRiT. The first row is the image of femur, the second row is
the image of brain lesions selected from ATLAS, and the third row is the lesion
structure added by ourselves. The red arrow in the image indicates the lesion
area, and the part circled by the red dotted oval is easier to distinguish the
reconstruction effect.
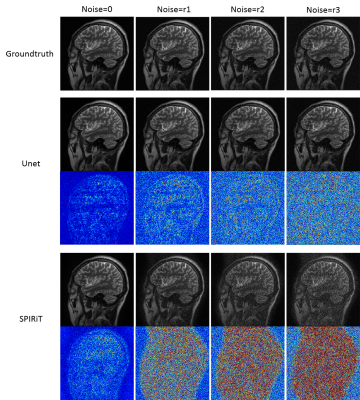
Fig .4 Add gaussian white noise to test
Unet and SPIRiT, and the noise level r3>r2>r1.
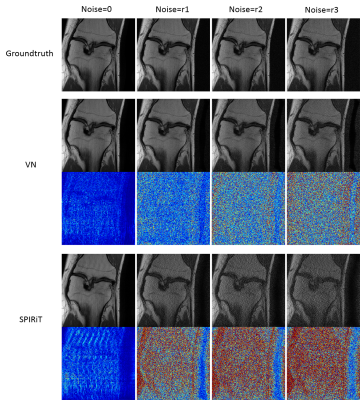
Fig .5 Add gaussian white noise to test VN
and SPIRiT, and the noise level r3>r2>r1.