3634
Learning reconstruction without ground-truth data: an unsupervised way for fast MR imaging1Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China, 2University at Buffalo, The State University of New York, Buffalo, Buffalo, NY, United States
Synopsis
Most deep learning methods for MR reconstruction heavily rely on the large number of training data pairs to achieve best performance. In this work, we introduce a simple but effective strategy to handle the situation where collecting lots of fully sampled rawdata is impractical. By defining a CS-based loss function, the deep networks can be trained without ground-truth images or full sampled data. In such an unsupervised way, the MR image can be reconstructed through the forward process of deep networks. This approach was evaluated on in vivo MR datasets and achieved superior performance than the conventional CS method.
Introduction
Deep learning (DL) has shown great promise at improving the quality of reconstructed MR image from highly undersampled k-space data1-6. Usually, it requires a large number of data pairs to train the network. The training data pairs usually consist of undersampled k-space data and the desired ground-truth image with respect to the corresponding full k-space, where the latter is used as the label to measure the training error. In practice, large number of full sampled k-space data are difficult to collect, and it is necessary to develop DL-based methods without ground-truth.In this work, we propose a novel deep learning reconstruction framework without access to full sampled data. Since there is no label in the training procedure, it is necessary to find an alternative loss function without label involved. Inspired by the compressed sensing (CS), where the optimal reconstruction is regarded as the solution of a L1-minimization problem, we use the objective function of CS problem to measure the distance between the reconstruction and ground-truth, thus this objective function serves as the loss function of the deep network. Experimental results show that the proposed method can achieve superior results compared to traditional CS method when reconstructing image from highly undersampled k-space data.
Theory
In supervised learning, the parameters of the network updates through network training to achieve the lowest error between the network output and the ground truth, where this learning direction is induced by the loss function. Loss function plays an important role in deep networks and different loss functions may result in different reconstructions.As the image reconstruction from undersampled k-space data is an ill-posed inverse problem, there is no unique solution. And among the potential solutions, CS aims to find the sparsest one. In general, the imaging model of CS-based methods7 can be written as$$\min_{m}\frac{1}{2} ‖Am-f‖_2^2+λ‖Ψm‖_1 (1)$$where the first term is the data consistency and the second term is the sparse prior. $$$Ψ$$$ is a sparse transform, such as wavelet transform or total variation, $$$m$$$ is the image to be reconstructed, $$$A$$$ is the encoding matrix, $$$f$$$ denotes the acquired k-space data.
CS searches the solution that minimize the objective function (1), and deep learning aims to train the network that could minimize the loss function. It comes naturally that the objective function of CS (1) can be used as the loss function of deep networks. In such scenario, no ground-truth or full sampled data are needed.
Methods
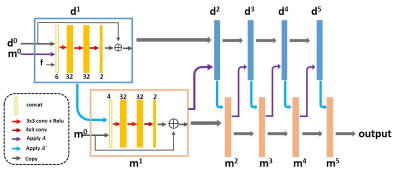
The network we use to present our approach is PD-net8, whose structure is shown in Fig 1. PD-net is an unrolling version of primal dual hybrid gradient (PDHG) algorithm that reconstructs the image by updating in k-space and image domain alternatively. The convolutions were all 3x3 pixel size, and the output of each CNN block has two channels representing the real and imaginary parts in the task of MR reconstruction. The loss function was defined as (1), where $$$λ=0.001$$$, $$$Ψ$$$ was chosen as total variation.Overall 2100 fully sampled multi-contrast data from 10 subjects with a 3T scanner (MAGNETOM Trio, SIEMENS AG, Erlangen, Germany) were collected and informed consent was obtained from the imaging object in compliance with the IRB policy. The fully sampled data was acquired by a 12-channel head coil with matrix size of 256×256 and adaptively combined to single-channel data and then retrospectively undersampled using Poisson disk sampling mask. 1400 undersampled data were used to train the networks. The proposed method was tested on the data from different 3T scanners (MAGNETOM Trio, SIEMENS AG, Erlgen, Germany; uMR 790, United Imaging Healthcare, Shanghai, China). The model was trained and evaluated on an Ubuntu 16.04 LTS (64-bit) operating system equipped with a Tesla TITAN Xp Graphics Processing Unit (GPU, 12GB memory) in the open framework Tensorflow with CUDA and CUDNN support.
Results
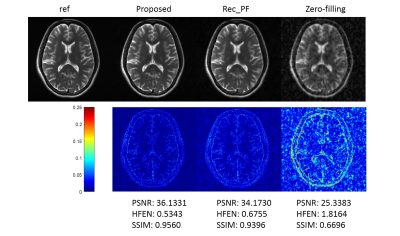
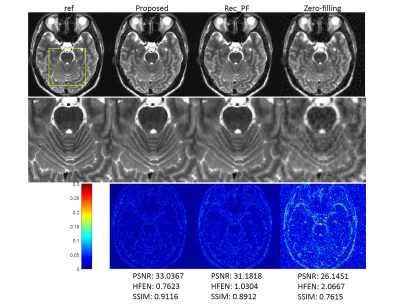
We compared the proposed method with the conventional CS-MRI method, Rec_PF9 with Bregman updating and the zero-filling reconstruction, the inverse Fourier transform of undersampled k-space data. Several similarity metrics, including PSNR, SSIM and HFEN, were used to compare the reconstruction results of different methods.Figure 2 illustrates the reconstructions of the different methods and the corresponding error maps with an acceleration factor of 6. Figure 3 shows the visual comparisons of different methods, where the zoom-in views demonstrate the superior performance of the proposed method.
The testing time of the proposed method for reconstructing an image of size 256*256 is 0.0710s, whereas 1.1688s for Rec_PF. The proposed method is much faster than CS method.
Conclusion
In this work, we proposed an effective unsupervised manner for deep learning MR reconstruction by designing a CS-based loss function. The effectiveness of the proposed strategy was validated on the in vivo MR data. The extension to other applications and more properties of the proposed framework will be explored in the future.Acknowledgements
This work was supported in part by the National Natural Science Foundation of China (U1805261). National Key R&D Program of China (2017YFC0108802) and the Strategic Priority Research Program of Chinese Academy of Sciences (XDB25000000).References
1. Wang S, Su Z, Ying L, et al. Accelerating magnetic resonance imaging via deep learning. ISBI 514-517 (2016)
2. Schlemper J, Caballero J, Hajnal J V, et al. A Deep Cascade of Convolutional Neural Networks for Dynamic MR Image Reconstruction. IEEE TMI 2018; 37(2): 491-503.
3. Yang Y, Sun J, Li H, Xu Z, ADMM-CSNet: A Deep Learning Approach for Image Compressive Sensing. IEEE Trans Pattern Anal Mach Intell, 2018.
4. Hammernik K, Klatzer T, Kobler E, et al. Learning a Variational Network for Reconstruction of Accelerated MRI Data. Magn Reson Med 2018, 79(6): 3055-3071.
5. Zhang J, Ghanem B. ISTA-Net: Interpretable Optimization-Inspired Deep Network for Image Compressive Sensing. CVPR 2018, 1828-1837.
6. Zhu B, Liu J Z, Cauley S F, et al. Image reconstruction by domain-transform manifold learning. Nature 2018; 555(7697): 487-492.
7. Lustig M, Donoho DL, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn Reson Med 2007; 58:1182–1195.
8. Cheng J, Wang H, Ying L, Liang D. Model Learning: Primal Dual Networks for Fast MR Imaging. MICCAI 2019
9. Yang J, Zhang Y, Yin W, et al. A Fast Alternating Direction Method for TVL1-L2 Signal Reconstruction From Partial Fourier Data. IEEE JSTSP 2010, 4(2): 288-297.
Figures