3633
Suppression of Artifact-Generating Echoes in Cine DENSE using Deep Learning1Biomedical Engineering, University of Virginia, Charlottesville, VA, United States
Synopsis
Cine displacement encoding with stimulated echoes (DENSE) is an accurate and reproducible method of strain imaging. The stimulated echo (STE), which carries the tissue displacement information in it’s phase, is simultaneously acquired with two artifact-generating echoes. A combination of phase-cycled acquisitions and through-plane dephasing are typically used to suppress the artifact-generating echoes. The limitations of these methods are longer acquisition times, susceptibility to breathing motion and the loss of signal-to-noise ratio due to intravoxel dephasing. To potentially overcome these limitations, the use of a deep convolutional neural network to suppress the undesired echoes from a single acquisition was investigated.
Introduction
Cine Displacement Encoding with Stimulated Echoes (DENSE) provides an accurate and reproducible measure of tissue displacement [1]. The DENSE signal consists of three echoes from three spin coherences: (1) the stimulated echo (STE), (2) the T1-relaxation echo (T1E) and (3) the conjugate stimulated echo (cSTE) [2]. Tissue displacement is encoded in the phase of the STE, while the two other echoes are undesired, generate artifacts and should be suppressed. A combination of phase-cycled acquisitions and the use of through-plane dephasing have been used to suppress the artifact-generating echoes for 2D cine DENSE. However, phase-cycling requires multiple data acquisitions and lengthens the scan time. In addition, it is a subtraction-based method and is susceptible to imperfect suppressions due to motion from imperfect breath-holds or free-breathing acquisitions. The use of through-plane dephasing increases intravoxel dephasing of the STE in deforming tissue, leading to a loss of STE signal-to-noise ratio (SNR). As an alternative, the echo suppression problem can be treated as a source separation problem. To potentially overcome the limitations of phase cycling and through-plane dephasing, we investigated the use of a deep convolutional neural network to suppress the undesired echoes and isolate the STE from a single acquisition.Methods
Breath-hold cine DENSE images of multiple anatomical views were acquired from 7 healthy subjects providing n=57 2D DENSE datasets with various displacement encoding frequencies; ke1=0.06 cyc/mm (n=13), ke2=0.03 cyc/mm (n=25), ke3=0.02 cyc/mm (n=6) and ke4=0.01 cyc/mm (n=13). A three-point phase cycling technique was used to suppress the T1E and the cSTE [3]. Three-point balanced displacement encoding was used to acquire data encoded for displacement along two orthogonal in-plane directions. Four spiral interleaves were acquired per cardiac phase with 2 interleaves/heartbeat resulting in 20 heartbeat breath-hold scans. The STEs were isolated by a linear combination of phase-cycled acquisitions. In order to create a training data set with a wider range of displacement encoding frequencies, a data augmentation method was developed where all three echoes were isolated with phase-cycling combinations. Then each echo was frequency re-modulated in k-space based on the following equation.$$S_{r,g}=\sum_{i=1}^{3} a_i S_{i,g}$$
$$ S_{i,g}=|S_{i}^{'}| e^{M_{E} \times C \times M_{D} \times \angle{S_{i}^{'}}} $$
$$ S_{i}^{'}=S_{i} e^{-i \theta_{i,b}} $$
Where Si=[si1, si2, si3]T is the vector of isolated ith echo along three displacement encoding directions, θi,b is the background phase associated with the ith echo, ME and MD are the three-point balanced encoding and decoding matrices [4] and the constant C is the displacement encoding frequency re-modulation constant. Then, the three frequency re-modulated (Si,g) echoes were added together by phase-cycling combination to simulate the raw DENSE data (Sr,g). The acquired DENSE images were augmented to cover displacement encoding frequencies ranging from 0.01 cyc/mm to 0.12 cyc/mm resulting in n=177 sets of DENSE images. The resulting dataset was divided into training (n=144), testing (n=17) and validation (n=16) sets. The k-spaces of all cardiac phases over the whole dataset were stacked for each set. A U-Net [5] with 5 encoding/decoding layers was trained where the input to the network was stacked k-spaces associated with one out of the three phase-cycling acquisitions and the ground-truth were the isolated STEs. A spatial transformation including translation and rotation in the image domain was used for data augmentation throughout the training. The validation set was used to find the optimal hyperparameters of the network. After the training, the test set was passed through the network to evaluate the performance of echo suppression. The Root Mean Squared Error was used on real and imaginary components of the images to quantify the performance of the network compared to the ground-truth.
Results
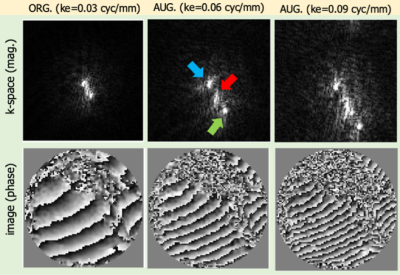
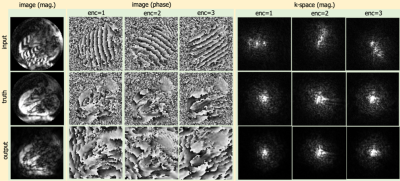
Figure 1 shows an example of data augmentation with displacement encoding frequency remodulation on a DENSE dataset acquired with ke=0.03 cyc/mm. The three echoes are more separated in k-space in the generated DENSE data with higher displacement encoding frequency. Figure 2 shows an example of echo suppression for images drawn from the test set for the three displacement-encoding directions, demonstrating that the trained network can suppress the T1E and cSTE and isolate the STE. The RMSE over entire test set was 0.16±0.07.Conclusion
This preliminary result shows that a deep convolutional neural network (a U-Net) is effective in isolating the STE and suppressing the undesired T1E and cSTE. This approach may be more effective than phase-cycling and through-plane dephasing, which have limitations. Future work will use more training data to improve the network and will demonstrate the value of this approach to echo suppression in the contexts of free-breathing 2D cine DENSE and accelerating 3D cine DENSE.Acknowledgements
No acknowledgement found.References
[1] D. Kim, W. D. Gilson, C. M. Kramer, and F. H. Epstein, “Myocardial tissue tracking with two-dimensional cine displacement-encoded MR imaging: development and initial evaluation,” Radiology, vol. 230, no. 3, pp. 862–871, 2004.
[2] X. Zhong, B. S. Spottiswoode, E. A. Cowart, W. D. Gilson, and F. H. Epstein, “Selective suppression of artifact-generating echoes in cine DENSE using through-plane dephasing,” Magn. Reson. Med. An Off. J. Int. Soc. Magn. Reson. Med., vol. 56, no. 5, pp. 1126–1131, 2006.
[3] J. Tsao and D. Laurent, “N-SPAMM for efficient displacement-encoded acquisition in myocardial tagging,” Proc. 13th Annu. Meet. ISMRM, Miami Beach, Florida, USA, vol. 42, no. 6, p. 273, 2005.
[4] X. Zhong, P. A. Helm, and F. H. Epstein, “Balanced Multipoint Displacement Encoding for DENSE,” vol. 988, pp. 981–988, 2009.
[5] O. Ronneberger, P. Fischer, and T. Brox, “U-net: Convolutional networks for biomedical image segmentation,” in International Conference on Medical image computing and computer-assisted intervention, 2015, pp. 234–241.
Figures