3632
Deep learning for undersampled spiral DENSE reconstruction1Imaging Sciences & Innovation, Geisinger, Danville, PA, United States, 2Medical & Health Physics, Geisinger, Danville, PA, United States, 3Radiology, Geisinger, Danville, PA, United States
Synopsis
Displacement Encoding with Stimulated Echoes (DENSE) is a powerful technique that has found great utility in accurately measuring cardiac tissue displacement. However, DENSE remains time-consuming to acquire, particularly for 3-dimensionally encoded or higher resolution schemes, and so methods to accelerate image acquisition are needed. Here, we apply the Deep Cascade of Convolutional Neural Networks (DCCNN) to the complex-valued, non-Cartesian data of DENSE to show that accelerated imaging via k-space undersampling is feasible using a deep learning-based reconstruction.
Introduction
Cine Displacement Encoding with Stimulated Echoes (DENSE) is a powerful but time-consuming technique used to obtain accurate and reproducible tissue displacement measurements1. Efforts to speed up DENSE have included an early switch to spiral trajectories, as well as parallel-imaging and compressed-sensing based approaches to reconstruction of undersampled data2. To reduce the number of phase cycles needed to suppress artifact-generating echoes, deep learning (DL) has been applied to DENSE3, but to date, these approaches have not been applied for undersampled image reconstruction. The Deep Cascade of Convolutional Neural Networks (DCCNN) technique makes use of concatenated CNNs, with maintenance of data fidelity through regular updates with the acquired k-space data4. It is extensible to non-Cartesian data in a straightforward manner and has the further advantage of maintaining the complex nature of the MRI data throughout the reconstruction pipeline, as well as handling multiple coils/contrasts. As DENSE is a spiral phase-contrast method, we sought to extend the DCCNN method to simulated undersampled data to explore the feasibility of accelerated DENSE imaging.Methods
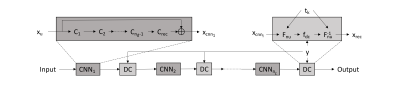
DENSE data from 5 volunteers obtained previously5 was used to generate training, validation, and test image sets. Between 10 and 14 slices from each volunteer were available, encoded in 3 dimensions over 31-39 cardiac phases. Two randomly selected slices (all cardiac phases) from each volunteer were reserved to serve as the validation and test data, respectively, resulting in total dataset sizes of 1678 images used for training, 171 for validation, and 171 for testing.The 2D DCCNN method was extended for non-Cartesian trajectories by replacing the Fourier transform operator at all applicable points with the nuFFT6 while maintaining appropriate scaling and mask operations (Fig. 1). The number of convolution layers (nd) in each network as well as the depth of the network (nc) were each set to 5. Pseudo raw data was generated by inverse gridding complex image data using a 12-interleaf, 6000-sample dual-density spiral trajectory. Undersampled data was generated by deleting every other interleaf. Trajectories were designed such that the center 33% of k-space remained fully-sampled after undersampling. X-, Y-, and Z- encoded images were treated as separate channels, and the resulting images were analyzed for general similarity to the reference using PSNR. Magnitude and phase SNR of the myocardium was recorded, and differences assessed by t-test. DENSEanalysis7 was used to interrogate the magnitude and variance of in-plane myocardial strain measurements generated by the reconstruction in comparison to the fully-sampled data.
Results
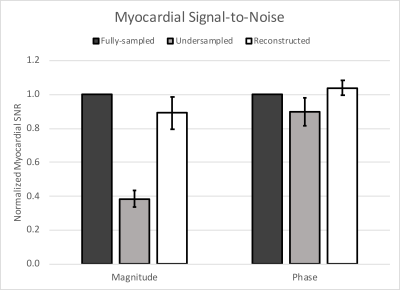
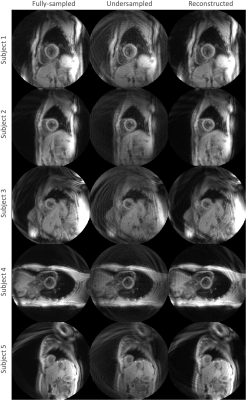
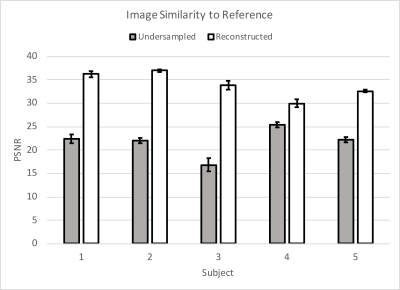
The myocardial magnitude SNR fell by 62% when undersampled, with nearly full recovery after reconstruction. Phase SNR was less affected; it was reduced by 10% when undersampled, but also recovered by the reconstruction (Fig. 2). Magnitude images from the sixth cardiac phase of each subject are shown in Figure 3. Fully-sampled, undersampled, and reconstructed images are shown left-to-right for each subject. The denoising performance varied with subject, with Subjects 1-3 experiencing a higher amount of denoising than 4 and 5, as measured by the PSNR of each image, compared to the fully-sampled reference image (Fig. 4). No differences in measured circumferential or radial strains were observed.Discussion
The reconstruction improved magnitude image appearance in every case; however, the relative gain varied across subjects. The relative improvement for Subject 4, in particular, was lower than in the others. It is notable that the orientation of the heart for Subject 4 led to a more axially-oriented slice than the others. This may highlight the importance of training on a wide variety of body types.The dual-density trajectories used for this study provide fully-sampled, low-resolution image data, contaminated with high frequency artifacts and noise. As the reconstruction can be viewed as a denoising problem, this type of trajectory proved sufficient to successfully reconstruct complex DENSE images from noisy undersampled data. The reconstruction failed when presented with undersampled constant-density spiral data (not shown) due to the resulting highly coherent artifacts. No difference in measured strains was observed at the current conservative acceleration factor. Future work might include optimizing the trajectory to minimize structured unsampling artifacts as higher acceleration rates are investigated.
Conclusion
Here, we have shown a DL approach with promise to reconstruct undersampled complex-valued spiral data, such as that used in DENSE. More investigation as to the type and number of anatomies needed for training, as well as the optimal pairing of trajectory design and DL algorithm, is needed.Acknowledgements
No acknowledgement found.References
1. Kim D, Gilson WD, Kramer CM, Epstein FH. Myocardial tissue tracking with two-dimensional cine displacement-encoded MR imaging: development and initial evaluation. Radiology. 2004 Mar;230(3):862-71.
2. Chen X, Yang Y, Cai X, Auger DA, Meyer CH, Salerno M, Epstein FH. Accelerated two-dimensional cine DENSE cardiovascular magnetic resonance using compressed sensing and parallel imaging. J Magn Reson Imaging. 2017 Sep;46(3):887-96.
3. Abdishektaei M, Feng X, Meyer CH, Epstein FH. DAS-Net: A generative adversarial net to suppress artifact-generating echoes in DENSE MRI. In ISMRM, Montreal, Canada, 2019, p1089.
4. Schlemper J, Caballero J, Hajnal JV, Price A, Rueckert D. A deep cascade of convolutional neural networks for MR image reconstruction. IEEE Trans Med Imaging. 2017;37(2):491-503.
5. Suever JD, Wehner GJ, Jing L, Powell DK, Hamlet SM, Grabau JD, Mojsejenko D, Andres KN, Haggerty CM, Fornwalt BK. Right ventricular strain, torsion, and dyssynchony in healthy subjects using 3D spiral cine DENSE magnetic resonance imaging. IEEE Trans Med Imaging. 2017 May;36(5):1076-85.
6. Lin J-M. Python non-uniform fast Fourier transform (PyNUFFT): An accelerated non-Cartesian MRI package on a heterogeneous platform (CPU/GPU)." J Imaging 2018;4(3):51.
7. Spottiswoode BS, Zhong X, Hess AT, Kramer CM, Meintjes EM, Mayosi BM, Epstein FH. Tracking myocardial motion from cine DENSE images using spatiotemporal phase unwrapping and temporal fitting. IEEE Trans Med imaging. 2007 Jan;26(1):15-30. Code available at: https://github.com/denseanalysis/denseanalysis
Figures