3631
Deep-learning reconstruction for 3D Delayed Myocardial Enhancement
Gaspar Delso1, Suryanarayanan Kaushik2, Graeme C McKinnon2, Daniel Lorenzatti3, Julián Vega3, Teresa M. Caralt3, Adelina Doltra3, José T. Ortiz-Pérez3, Rosario J. Perea3, Susanna Prat3, Marta Sitges3, and Martin A. Janich4
1ASL MR, GE Healthcare, Barcelona, Spain, 2GE Healthcare, Waukesha, WI, United States, 3Hospital Clínic de Barcelona, Barcelona, Spain, 4GE Healthcare, Munich, Germany
1ASL MR, GE Healthcare, Barcelona, Spain, 2GE Healthcare, Waukesha, WI, United States, 3Hospital Clínic de Barcelona, Barcelona, Spain, 4GE Healthcare, Munich, Germany
Synopsis
We present the evaluation results of a novel Deep Learning reconstruction framework, applied to clinical Delayed Myocardial Enhancement datasets acquired with a 3D inversion-recovery T1-weighted gradient echo sequence.
INTRODUCTION:
Delayed Enhancement (DE) MRI is a valuable imaging technique for the identification of a number of ischemic and non-ischemic (e.g. myocarditis, idiopathic dilated cardiomyopathy, amyloidosis) cardiac aetiologies. DE-MRI is based on the acquisition of T1-weighted images after an initial gadolinium contrast injection is allowed a 10- to 20-minute washout time1.Three-dimensional (3D) imaging offers increased resolution and full heart coverage, improving the understanding of complex pathological patterns. A relatively long acquisition time, strongly dependent on the cardiorespiratory condition of the patient, is required for this purpose. It follows that there is an intrinsic trade-off with the achievable image quality, potentially limiting the ability to identify low-contrast pathological uptake.
Deep learning (DL) reconstruction methods have recently been shown to be an effective way to incorporate prior knowledge in the MR reconstruction pipeline, improving regularization of the inverse problem solution2. We postulate that the application of DL techniques can effectively overcome the trade-off between reconstructed image quality and acquisition time in 3D-DE imaging, opening a way towards accelerated acquisition methods.
In this study we validate a novel three-dimensional Deep Learning reconstruction framework on Delayed Myocardial Enhancement datasets of clinical subjects.
METHODS:
A set of 14 cases (9 male, 5 female, age 59±11 years, weight 78±13 kg) were selected for this retrospective study. All subjects had undergone a clinically indicated contrast-enhanced cardiac examination on a GE Architect 3T MRI at Hospital Clínic of Barcelona.The standard clinical acquisition protocol was followed by a 3D Delayed Enhancement acquisition using a prototype pulse sequence. The sequence settings were: SPGR readout with IR preparation, fat suppression, spatial saturation, respiratory navigator, 2x acceleration (ARC), 40x40cm FOV, ST 1.4-2.4mm, matrix 2802-3202, FA 20deg, BW 62.5 kHz, TE 2.1±0.1ms, TI 269±36ms (based on a CINE IR scout), trigger time 585±107ms. The raw data of these acquisitions was anonymized and exported for further analysis.
A prototype 3D DL Recon algorithm, implemented using GE Healthcare’s Orchestra libraries, was used to retrospectively reconstruct all the exported datasets. The reconstruction incorporated a 3D model trained on a database of over 700 data sets to reconstruct high quality 3D images with a tuneable noise reduction factor. A reference reconstruction was also performed with a 3D Cartesian pipeline mimicking that implemented in the scanner.
Standard deviation measurements were performed on a manually defined region of interest in the atrial blood pool, as a surrogate for overall image noise. The Structural Similarity Index (SSIM) was used to compare the DL reconstruction results with those of the standard reconstruction, as an estimate of perceptually relevant structure preservation.
All reconstructions were screened by two board-certified cardiologists with experience in MRI reading, in order to identify any clinically relevant alterations in morphology or pathological uptake.
RESULTS:
All datasets were successfully reconstructed with the new 3D DL Recon. From a qualitative point of view, the new method consistently resulted in lower noise images without any noticeable loss of structural information or spatial resolution (figures 1 and 2).These results were confirmed by the quantitative analysis (figure 3). For a given level of blood pool standard deviation (i.e. image noise), the Deep Learning approach yielded up to 10% better structural similarity to the reference image, compared to conventional anisotropic filtering3. ROI analysis showed an average coefficient of variation in the blood pool of 0.08±0.02 with the standard reconstruction and 0.05±0.02 with the deep learning, corresponding to a 35%±12% reduction of standard deviation.
Expert screening didn’t reveal any clinically relevant alterations of cardiac morphology or delayed enhancement patterns. The overall noise reduction was initially perceived as a loss of feature sharpness. However, this notion was disproved upon closer examination of specific anatomical landmarks (e.g. pulmonary and coronary arteries). The improved uniformity of myocardial regions was reported to reduce the likelihood of false positive diagnosis of fibrotic tissue, although this claim has not been quantified.
DISCUSSION:
Deep Learning results were found to provide the best trade-off between blood pool standard deviation and structural similarity index (with respect to the standard reconstruction results), as compared to off-the-shelf post reconstruction denoising techniques. This consistent improvement of image quality, together with the fact that no morphological alterations of diagnostic relevance were found by the expert screening, suggests that the new method can be reliably introduced into clinical practice.A limitation of the present study is the relatively small number of cases evaluated. Additionally, all cases were acquired at a site with considerable expertise in 3D Delayed Enhancement cardiac imaging. While no pre-selection of the cases was done, it would still be advisable to extend the study to a larger population of more heterogeneous image quality.
CONCLUSION:
The results obtained with a new 3D Deep Learning reconstruction on Delayed Enhancement cardiac data indicate that this method can consistently improve image quality in a clinical setting.Acknowledgements
No acknowledgement found.References
1. Dulce, M. C. et al. MR imaging of the myocardium using nonionic contrast medium: signal-intensity changes in patients with subacute myocardial infarction. AJR Am J Roentgenol 160, 963–970 (1993).
2. Hyun, C. M., Kim, H. P., Lee, S. M., Lee, S. & Seo, J. K. Deep learning for undersampled MRI reconstruction. Phys. Med. Biol. 63, 135007 (2018).
3. Gerig, G., Kubler, O., Kikinis, R. & Jolesz, F. A. Nonlinear anisotropic filtering of MRI data. IEEE Transactions on Medical Imaging 11, 221–232 (1992).
Figures
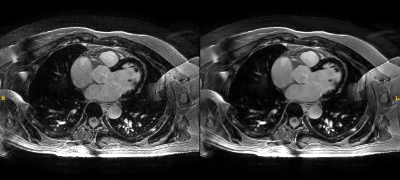
Figure 1.- Axial view
of a 3D Delayed Enhancement series, reconstructed with the standard (left) and
Deep Learning method (right). Notice the subtle septal uptake.
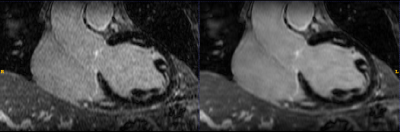
Figure 2.- Detail of
coronal view of a 3D Delayed Enhancement series, reconstructed with the
standard (left) and Deep Learning method (right).
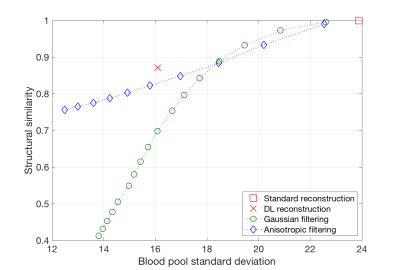
Figure 3.- Structural
similarity to the standard reconstruction method, compared to Gaussian and
anisotropic post-reconstruction filtering results (over a range of their
respective parameter settings). Notice how Deep Learning reconstruction yields
the best compromise between noise reduction and structural similarity.