3625
Simultaneous Brain Anatomical and Arterial Imaging by 3T MRI: Reconstruction Based on a Generative Adversarial Network1Sinovation Ventures AI Institute, Beijing, China, 2QuantMind, Beijing, China, 3MR Collaboration, Siemens Healthcare, Beijing, China, 4State Key Laboratory of Brain and Cognitive Science, Beijing MRI Center for Brain Research, Institute of Biophysics, Chinese Academy of Sciences, Beijing, China, 5University of Chinese Academy of Sciences, Beijing, China, 6Beijing Institute for Brain Disorders, Beijing, China
Synopsis
This study investigates a reconstruction method designed to generate 7T magnetic resonance images (MRI) from 3T MRI based on a generative adversarial network. Compared with current reconstruction methods, this method can simultaneously reconstruct well-defined anatomical details and salient blood vessels. This reconstruction method may be useful for increasing the efficiency of brain MRI examinations.
Introduction
Based on the field intensity of magnetic scanning, magnetic resonance imaging (MRI) can be generally classified into three groups: 1.5T, 3T, and 7T. Because of its higher resolution and higher signal-to-noise ratio (SNR), 7T MRI can provide more accurate imaging of brain metabolic processes and anatomical details compared to routine 1.5T and 3T MRI. This may be beneficial in the diagnosis of brain diseases such as Alzheimer’s disease, Parkinson's disease, and other dementia.1 However, 7T MRI scanners are expensive and therefore less commonly used in the clinic. Thus, it may be worthwhile to reconstruct high-field MRI from low-field MRI. Recent studies proposed 7T reconstruction as a super-resolution target 2-4 to obtain 7T-like MRI with well-defined anatomical details. However, blood vessels, which generate higher signals in 7T than in 3T MRI, generally cannot be reconstructed. We proposed a generative adversarial network (GAN)-based deep learning algorithm designed to improve the images acquired by 3T MRI, to obtain reconstructed 7T MRI images with distinct anatomical details and salient blood vessels.Methods
The FocalGAN model was based on a GAN 5 and is designed to capture the distribution of a 7T MRI target domain and transfer the 3T MRI source domain to the target domain using adversarial training. The main features of the target domain are clearer anatomical details and salient blood vessels. Thus, through adversarial learning, anatomical details and high signal intensity blood vessels can be generated. However, because of a GANs' instability and randomness, the position of blood vessels tends to be inaccurate and the visual quality of the results is poor. Therefore, we added focal loss 6 to constrain the generative ability of the model and to help focus the model on the regions required to be recovered. Additionally, perceptual loss 7 was added, which reduces feature differences between reconstructed results and 7T MRI, to further improve the visual performance of results and enable the generated results appear more like 7T MRI on the semantic level.Network Architecture
FocalGAN consisted of a generator and a discriminator:
1.The generator was a 3D U-Net-like 8 network, using 4 times down-sampling and 4 times up-sampling with short connections. 3T MRI was used as input and the output generated were reconstructed results. The entire network optimized adversarial loss, focal loss, and perceptual loss.
2.The discriminator was a 5-layer 3D classification network. Reconstructed results or 7T MRI were used as input; the output corresponded to classification results. The discriminator was used to distinguish between real data (7T MRI) and generated data. The entire network optimized only adversarial loss.
Data Acquisition
Paired 3T and 7T T1-weighted magnetization-prepared 180 degrees radio-frequency pulses and rapid gradient-echo (MP RAGE) MR images were acquired from 20 patients scanned using a Siemens Magnetom TimTrio 3T MR scanner and Siemens Magnetom 7T MR Scanner. 3T and 7T MRI were first resampled to a fixed brain template, the Multimodal Brain Tumor Image Segmentation Benchmark (BRATS) template, 9 whose pixel spacing is 1mm. 7T MRI was registered to the 3T MRI using affine transformation. The brain template was registered to the 3T to acquire brain skull masks using nonlinear transformation using symmetric normalization with cross-correlation (SyNCC ) in Advanced Normalization Tools (ANTs). Brain-skull-removed 3T MRI and 7T MRI were registered to the brain template using affine transformation. Finally, N4 bias field correction was applied to the data. The matrix sizes of the prepared 3T and 7T MRI were 155, 240, and 240. Eighteen pairs were used for training, and the remaining two were used for testing. FocalGAN was trained on a NVIDIA TITAN X using Adam in the TensorFlow2.0 library.
Discussion
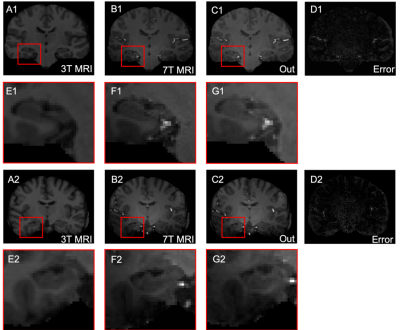
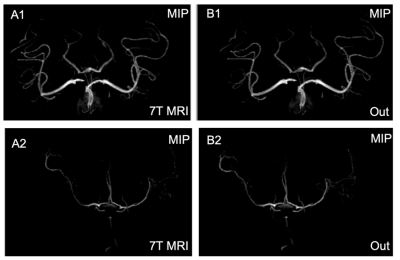
As shown in Figure 1, the hippocampus was highlighted in 7T MRI; the results demonstrate greater contrast and more well-defined edges compared with that seen on 3T MRI. The error maps show that the results can provide lower mean square error (MSE), as well as generate similar images to those generated by 7T MRI. The MIP images shown in Figure 2 demonstrate that FocalGAN can reconstruct vascular structures that appear only slightly different from the source 7T MRI.Conclusion
The FocalGAN method was proposed to reconstruct 7T MRI from 3T MRI. Compared with existing reconstruction methods, this method not only can generate well-defined anatomical details but can also reconstruct vascular structures. Thus, this method may be valuable in clinical diagnosis.Acknowledgements
National Nature Science Foundation of China grant: 61473296,81871350References
1. Springer E, Dymerska B, Cardoso P L, et al. Comparison of routine brain imaging at 3 T and 7 T[J]. Investigative radiology, 2016, 51(8): 469.
2. Qu L, Wang S, Yap P T, et al. Wavelet-based Semi-supervised Adversarial Learning for Synthesizing Realistic 7T from 3T MRI[C]//International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Cham, 2019: 786-794.
3. Dual-domain cascaded regression for synthesizing 7T from 3T MRI
4. Zhang Y, Cheng J Z, Xiang L, et al. Dual-domain cascaded regression for synthesizing 7T from 3T MRI[C]//International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Cham, 2018: 410-417.
5. Goodfellow I, Pouget-Abadie J, Mirza M, et al. Generative adversarial nets[C]//Advances in neural information processing systems. 2014: 2672-2680.
6. Lin T Y, Goyal P, Girshick R, et al. Focal loss for dense object detection[C]//Proceedings of the IEEE international conference on computer vision. 2017: 2980-2988.
7. Ledig C, Theis L, Huszár F, et al. Photo-realistic single image super-resolution using a generative adversarial network[C]//Proceedings of the IEEE conference on computer vision and pattern recognition. 2017: 4681-4690.
8. Çiçek Ö, Abdulkadir A, Lienkamp S S, et al. 3D U-Net: learning dense volumetric segmentation from sparse annotation[C]//International conference on medical image computing and computer-assisted intervention. Springer, Cham, 2016: 424-432.
9. Rohlfing T, Zahr N M, Sullivan E V, et al. The SRI24 multichannel atlas of normal adult human brain structure[J]. Human brain mapping, 2010, 31(5): 798-819.
Figures