3613
Super-resolution MRI using deep convolutional neural network for adaptive MR-guided radiotherapy: a pilot study1Medical Physics & Research Department, Hong Kong Sanatorium & Hospital, Hong Kong, China, 2Department of Biomedical Engineering, Department of Electrical Engineering, The State University of New York at Buffalo, Buffalo, NY, United States
Synopsis
MR-guided radiotherapy (MRgRT) is creating new perspectives towards an individualized precise radiation therapy solution. However, spatial resolution of fractional MRI can be much restricted, in order to shorten scan time, by patient tolerance of immobilization, intra-fractional anatomical motion and complicated MRgRT workflow. We hypothesized that the quality of low-resolution daily MRI could be greatly restored to generate super-resolution MRI, whose quality should be comparable of high-resolution planning MRI, by applying deep learning techniques. In this study, we aimed to investigate the feasibility of deep learning super-resolution MRI generation in the head-and-neck for adaptive MRgRT purpose.
Introduction:
MR-guided radiotherapy (MRgRT) is creating new perspectives towards an individualized precise radiation therapy solution by directly visualizing tumor, surrounding tissues and their motions for treatment adaptation and radiation delivery guidance. However, the spatial resolution of fractional (or daily) MRI can be much restricted, to shorten the scan time, by patient tolerance of immobilization, intra-fractional anatomical motion, and complicated MRgRT workflow. This potentially leads to compromised tissue delineation, image registration and/or treatment adaptation. We hypothesized that the quality of low-resolution daily MRI could be greatly restored to generate super-resolution MRI, whose quality should be comparable to high-resolution planning MRI, by applying deep learning techniques, like convolutional neural network (CNN) and recurrent neural networks (RNN)1-8. In this study, we aimed to test the feasibility of deep learning super-resolution MRI generation in the head-and-neck for adaptive MRgRT purpose.Material and Methods:
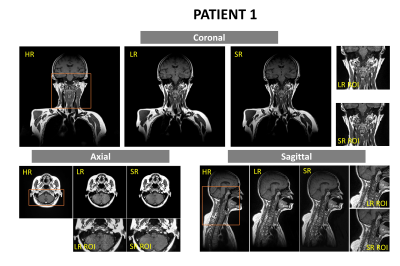
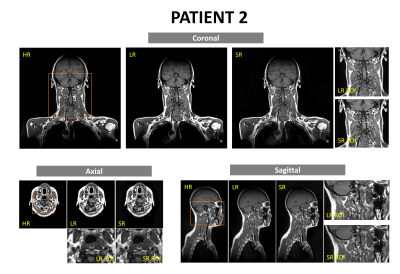
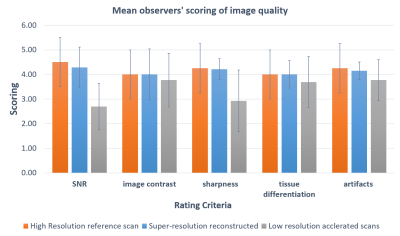
18 healthy volunteers and 2 head-and-neck cancer (HNC) patients were recruited, and scanned on a 1.5-Tesla MR-simulator (Aera, Siemens Healthineers, Erlangen, Germany) in the immobilized RT treatment position. 18 volunteers received 183 paired MRI scans. Each paired scan included a high-resolution T1w SPACE acquisition (FOV = 470× 470 × 269 mm3 and matrix size = 448 × 448 × 256; TR/TE = 420/7.2 ms, echo train length (ETL)= 40, GRAPPA factor = 3; RBW= 657Hz/pixel, isotropic voxel size of 1.05mm, duration=5min) using planning MRI protocol and a highly accelerated low-resolution acquisition (same TE/TR/ETL/RBW, GRAPPA factor = 9, voxel size = 1.4x1.4x1.4mm3, duration=86s) using daily MRI protocol9,10. For patients, each received one high-resolution planning MRI and a series of low-resolution daily MRI (3 fractions for patient 1 and 11 fractions for patient 2). 185 paired high-resolution and low-resolution image sets (including 183 volunteer sets and 2 patient sets), were used as training data. The patients’ unpaired low-resolution image sets (2 for patient 1 and 10 for patient 2) were used to generate super-resolution MRI (the same resolution as planning MRI) for validation and testing. No image registration between paired low-resolution and high-resolution image sets was needed by taking advantage of the same scan position under immobilization. A previously proposed deep CNN (DCNN) network was modified11,12. The nonlinear relationship Θ between the low-resolution and high-resolution images, expressed as $$$[{I_{HR}} = F({I_{LR}};\Theta )]$$$, was learned through minimizing the loss function between the network prediction and the corresponding ground truth data by residual learning algorithm, or mathematically $$$\arg {\min _\Theta }\frac{1}{n}\sum\nolimits_{i = 1}^n {\left\| {F({I_{LR}};\Theta ) - {I_{HR}}} \right\|_2^2}$$$, where n is the number of training datasets. 15 weighted CNN layers were used for training and testing. Except for the last one, each layer included 64 filters. A weight decay of 0.0001 and a learning rate of 0.0001 for 1.4M iterations with ReLU as the activation function were used. Two medical physicists blindly evaluated image quality on a 5-point scale (1 poor, 2 fair, 3 moderate, 4 good, 5 excellent), based on SNR, image contrast, sharpness, tissue differentiation, and artifacts. Cohen kappa analysis ($$$\kappa $$$) and the Wilcoxon signed-rank test (significant level p=0.05) was used to assess the rating result.Results
The training of the DCNN network took about 24h on a workstation (CPU i9-7980XE; Memory 64 GB; GPU 2x NVIDIA GTX 1080Ti). The testing only took seconds. The super-resolution images removed most noise and restored fine structure information of important small organs-at-risks (OARs) (Fig. 1). Two medical physicists showed excellent inter-observer scoring agreements ($$$\kappa > 0.8$$$). The rating of the super-resolution images was slightly lower than the high-resolution images but without significant difference (p =0.0791). Super-resolution images were rated significantly better than low-resolution images. (p<0.0001).Discussion and Conclusion
In this study, we investigated the feasibility of applying deep convolutional neural network to recover 3D high-resolution images from low-resolution images for MRgRT in the head-and-neck. Preliminary results show that the super-resolution images using DCNN were able to remove image noises and restore the spatial details of fine OAR structures which were missed by the highly accelerated MRI scans. Although the network training was mainly based on volunteer data, the trained network successfully applied to patient super-resolution MRI generation. The generation of super-resolution MRI holds the nominal same spatial resolution as planning MRI, but with 72% shorter acquisition time, so it could potentially facilitate efficient and accurate MRgRT workflow. The main limitation of this study is that our experiments were based on only two patients’ data and mixed healthy volunteers, as well as the small sample size. Further validation on the robustness of the proposed method based on larger samples of real HN cancer patient data is warranted.Acknowledgements
This study was approved by the Institutional Research Ethics Committee (REC-2019-11)References
[1] S. Wang, Z. Su, L. Ying, X. Peng, S. Zhu, F. Liang, D. Feng, D. Liang, “Accelerating magnetic resonance imaging via deep learning,” IEEE 14th International Symposium on Biomedical Imaging (ISBI), pp. 514-517, Apr. 2016.
[2] S. Wang, N. Huang, T. Zhao, Y. Yang, L. Ying, D. Liang, “1D Partial Fourier Parallel MR imaging with deep convolutional neural network,” Proceedings of International Society of Magnetic Resonance in Medicine Scientific Meeting, 2017.
[3] D. Lee, J. Yoo and J. C. Ye, “Deep residual learning for compressed sensing MRI,” IEEE 14th International Symposium on Biomedical Imaging (ISBI), Melbourne, VIC, pp. 15-18, Apr. 2017.
[4] Chang Min Hyun et al, “Deep learning for undersampled MRI reconstruction,” Phys. Med. Biol. 63 135007, 2018
[5] S. Wang, T. Zhao, N. Huang, S. Tan, Y. Liu, L. Ying, and D. Liang, “Feasibility of Multi-contrast MR imaging via deep learning,” Proceedings of International Society of Magnetic Resonance in Medicine Scientific Meeting, 2017
[6] B Zhu, JZ Liu, SF Cauley, BR Rosen, MS Rosen. Image reconstruction by domain-transform manifold learning. Nature. 2018 Mar;555(7697):487.
[7] K. Hammernik, T. Klatzer, E. Kobler, M. P. Recht, D. K.Sodickson, T. Pock, and F. Knoll, “Learning a variational network for reconstruction of accelerated MRI data,”Magnetic Resonance in Medicine, vol. 79, no. 6, pp.3055–3071, 2018.
[8] K. H. Jin, M. T. McCann, E. Froustey and M. Unser, “Deep Convolutional Neural Network for Inverse Problems in Imaging,” in IEEE Transactions on Image Processing, vol. 26, no. 9, pp. 4509-4522, Sept. 2017.
[9] Y. Zhou, J. Yuan, OL. Wong, WWK. Fung, KF. Cheng, KY. Cheung, SK Yu. Assessment of positional reproducibility in the head and neck on a 1.5-T MR simulator for an offline MR-guided radiotherapy solution. Quant Imaging Med Surg 2018;8:925-35.
[10] Y. Zhou, OL. Wong, KY. Cheung, SK. Yu, J. Yuan. A pilot study of highly accelerated 3D MRI in the head and neck position verification for MR-guided radiotherapy. Quant Imaging Med Surg 2019;9(7):1255-1269.
[11] H. Li, C. Zhang, U. Nakarmi, et al, “MR knee image reconstruction using very deep convolutional neural networks”, ISMRM workshop on machine learning, #59, Pacific Grove, USA, 2018.
[12] P. Zhou, C. Zhang, H. Li, et al, “Deep MRI reconstruction without ground truth for training”, ISMRM, Montreal, Quebec, Canada, #4668, 2019.
Figures