3593
A direct MR image reconstruction from k-space via End-To-End reconstruction network using recurrent neural network (ETER-net)
Changheun Oh1, yeji han2, and HyunWook Park1
1Electrical Engineering, Korea Advanced Institute of Science and Technology, Daejeon, Korea, Republic of, 2Biomedical engineering, Gachon University, Incheon, Korea, Republic of
1Electrical Engineering, Korea Advanced Institute of Science and Technology, Daejeon, Korea, Republic of, 2Biomedical engineering, Gachon University, Incheon, Korea, Republic of
Synopsis
In this work, we propose a novel neural network architecture named ‘ETER-net’ as a unified solution to reconstruct an MR image directly from k-space data. The proposed image reconstruction network can be applied to k-space data that are acquired with various scanning trajectories and multi or single-channel RF coils. It also can be used for semi-supervised domain adaptation. To evaluate the performance of the proposed method, it was applied to brain MR data obtained from a 3T MRI scanner with Cartesian and radial trajectories.
Introduction
With recent advances in deep learning research, many approaches have been introduced for deep learning based image reconstruction, which mostly focused on CNN-based models for image domain datasets. However, the sensor domain data obtained from the MR scanner is not in the image domain but in the k-space domain. Moreover, the k-space data is undersampled in many cases to reduce imaging time. The proposed deep learning based end-to-end method aims to solve global transformation (from the k-space domain to the image domain) and to generate a de-aliased the image from undersampled input k-space data. Recently, AUTOMAP was introduced as an end-to-end reconstruction method which transformed an image from input k-space data[1], but it was difficult to utilize AUTOMAP to reconstruct large sized images due to high memory requirements of its fully connected layers. In addition, MRI is often performed with multi-channel RF coils, generating multi-channel datasets, but there are only a few studies using multi-channel MR data[2,3]. This study proposes a deep learning based image reconstruction method that generates images from input k-space data by a bidirectional RNN-based end-to-end network named ‘ETER-net’[4]. The proposed method can be used for 256×256 size images and for multi-channel MR data acquired with various sampling trajectories. In addition, our experiment result shows that the proposed network can be used for semi-supervised domain adaptation, e.g. the network trained by large T2w dataset and small T1w dataset can be used for T1w image reconstruction.Methods
As presented in Fig. 1, the proposed network, ETER-net, consists of two parts: a domain transform network and a refinement network. The domain transform network consists of two bidirectional RNN layers, which transforms the input k-space data into image-like features. Each bidirectional recurrent layer performs global transformation and de-aliasing. In the refinement network, the features generated by the domain transform network is refined and merged into a final image. The refinement network can be freely designed for specific needs but we adopted the dual frame U-net (DFU) in this study[5]. To experiment with various sampling trajectories, we used three different types of sampling trajectories; Cartesian regular sampling with a reduction factor of R=4, Cartesian regular sampling with R=4 and additional center lines (9 ACS lines, Reff=3.6), and radial sampling (R=4). In-vivo experiments were performed using a 3.0T MRI system (Siemens Magnetom Verio, Erlangen, Germany) to acquire datasets with the following parameters. Cartesian : the number of images is 1500 for training and 60 for testing, matrix size = 216 (frequency encoding) × 216 (phase encoding) × 8 (RF coil channels), slice thickness = 5 mm, echo train length = 18 for the fast spin-echo sequence and TR/TE = 5000/90 ms (T2w), 558/10 ms (T1w). Radial : the number of images is 2500 for training and 48 for testing, matrix size = 256 (frequency encoding) × 400 (view angles) × 4 (RF coil), slice thickness = 5 mm, echo train length = 8 and TR/TE = 5000/52 ms.The proposed method was compared with previous methods, such as algorithmic methods (GRAPPA, Radon) and deep learning based methods (DFU, DAGAN)[6]. To verify that the proposed method can be used for semi-supervised domain adaptation, we applied T1w k-space data to the networks trained by three different training sets (T1w dataset (1216 samples), T2w dataset (4590 samples), and a combination of large amount of T2w(4590 samples) and small amount of T1w (300 samples) datasets). The test set was comprised of 60 T1w k-space data.Results & Discussion
We used the normalized mean squared error (nMSE) and the structural similarity (SSIM) for quantitative evaluation . Fig. 2 illustrates sample images and the corresponding error maps acquired with three different trajectories: regular sampling (1st row), regular sampling with additional center lines (2nd row), and radial sampling (3rd row). The quantitative evaluation results are presented in Table 1. As shown in Fig. 2, the reconstructed images from previous methods have reconstruction errors or aliasing artifacts, while images reconstructed by the proposed network have negligible reconstruction errors. For semi-supervised domain adaptation, different combination of T1w and T2w datasets were used for training of the proposed network. For reconstruction of a T1w image, utilizing the T1w k-space dataset for training would naturally produce the best results, and utilizing the T2w dataset for training would produce images with contrast differences. As demonstrated in Fig. 3, the network trained by T2w datasets generated images with a T2w-like contrasts and aliasing artifacts. However, the network trained by a large portion of T2w dataset and a small portion of T1w dataset, where the ratio of T2w dataset and T1w dataset was is 15:1, provided images having T1w contrasts. Moreover, the error maps showed that utilizing a combination of T1w and T2w generated better images than utilizing the target domain (domain of testing dataset, i.e., T1w) dataset only, probably due to proper generalization that might have been learned from general characteristics in both domains and the abundant amount of datasets. In other words, we can apply the proposed network to target domain dataset if we train the network with an abundant source domain (domain of abundant training dataset, i.e., T2w) dataset and a small amount of target domain dataset.Acknowledgements
This work was partially supported by the Brain Research Program through the National Research Foundation of Korea (NRF) funded by the Korea government (MSIT) (NRF-2017M3C7A1047228) and Institute for Information & communications Technology Planning & Evaluation (IITP) grant funded by the Korea government (MSIT) (No.2017-0-01779, A machine learning and statistical inference framework for explainable artificial intelligence) .References
1. Zhu, B., Liu, J. Z., Cauley, S. F., Rosen, B. R., & Rosen, M. S. (2018). Image reconstruction by domain-transform manifold learning. Nature, 555(7697), 487.2. Kwon, K., Kim, D., & Park, H. (2017). A parallel MR imaging method using multilayer perceptron. Medical physics, 44(12), 6209-6224.3. Duan, J., Schlemper, J., Qin, C., Ouyang, C., Bai, W., Biffi, C., ... & Rueckert, D. (2019, October). VS-Net: Variable splitting network for accelerated parallel MRI reconstruction. In International Conference on Medical Image Computing and Computer-Assisted Intervention (pp. 713-722). Springer, Cham.4. Oh, C., Kim, D., Chung, J. Y., Han, Y., & Park, H. (2018, September). ETER-net: End to end MR image reconstruction using recurrent neural network. In International Workshop on Machine Learning for Medical Image Reconstruction (pp. 12-20). Springer, Cham.5. Han, Y., & Ye, J. C. (2018). Framing U-Net via deep convolutional framelets: Application to sparse-view CT. IEEE transactions on medical imaging, 37(6), 1418-1429.6. Yang, G., Yu, S., Dong, H., Slabaugh, G., Dragotti, P. L., Ye, X., ... & Firmin, D. (2017). DAGAN: deep de-aliasing generative adversarial networks for fast compressed sensing MRI reconstruction. IEEE transactions on medical imaging, 37(6), 1310-1321.Figures
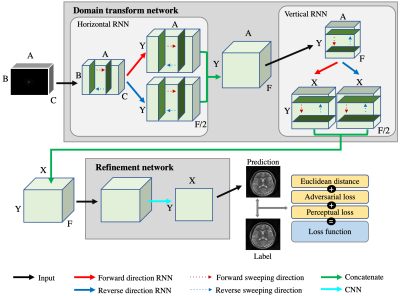
A diagram of the proposed End-To-End reconstruction network using recurrent neural network (ETER-net)
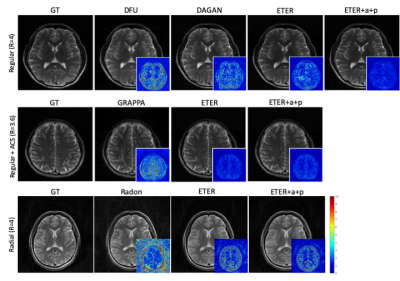
Reconstructed images from previous and the proposed methods (Intensity of the error maps is amplified by 10)
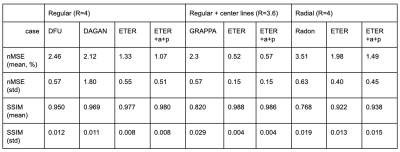
Table 1. Evaluation results of the previous method and the proposed method with three different sampling trajectories: regular sampling (R=4), regular sampling with additional center lines (R=3.6), and radial sampling (R=4). +a : adversarial loss based on patch-GAN, +p : perceptual loss based on VGG network.
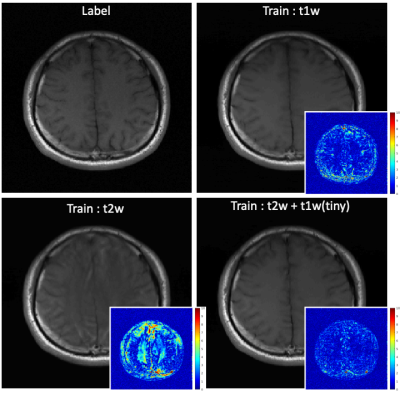
Experimental results of semi-supervised domain adaptation for image reconstruction using the proposed method