3590
Model-Free Deep MRI Reconstruction: A Robustness Study1Medical Physics, Memorial Sloan Kettering Cancer Center, New York, NY, United States, 2Radiology, Memorial Sloan Kettering Cancer Center, New York, NY, United States
Synopsis
Speed is often claimed as a key advantage of deep learning (DL) for undersampled parallel MRI reconstruction. However, leading DL methods require repeated application of the MR acquisition model and its adjoint, just as in conventional iterative methods. This work investigates the feasibility and robustness of model-free DL reconstruction, which has the potential to be much faster. Results in varied patient cases of increasing pathological rarity demonstrate that while model-free DL can reasonably reconstruct anatomies similar to those seen in training, performance can degrade drastically in more challenging situations.
Introduction
There is growing interest in applying deep learning (DL) for undersampled parallel MRI reconstruction. A key advantage often claimed of DL over conventional iterative methods is reconstruction speed1. However, the only DL approach that to our knowledge has studied generalizability to pathologies unseen in training2 requires repeated application of the MR acquisition model and its adjoint, just as in iterative methods. In contrast, model-free DL reconstruction has the potential to be much faster. Here we investigate the feasibility of model-free DL reconstruction, using a neural network that operates entirely in the image domain by mapping aliased multicoil images to an unaliased image. The model-free DL approach is tested in challenging patient cases with a variety of brain tumors.Methods
Data acquisition. 3D BRAVO (T1-weighted MRI) raw datasets were acquired in 48 volunteers and patients, across 4 different GE Healthcare 3T scanners (2 MR750w, 1 MR750, and 1 Premier) at our institution. 3D k-space was fully-sampled in each acquisition to obtain a reference and to enable the testing of different acceleration factors through retrospective undersampling. Acquisition matrix sizes ranged from 256x206x168 to 256x260x336.Data preparation. Each dataset was retrospectively undersampled by an overall factor of about 4 along the two phase-encoding dimensions; coil-compressed3 to reduce the influence of coil geometry in training; and then Fourier-transformed coil-by-coil, yielding aliased virtual coil images. The results shown in this abstract correspond to a regular 2x2 sampling pattern that matches the pattern of a conventional ARC parallel imaging acquisition.
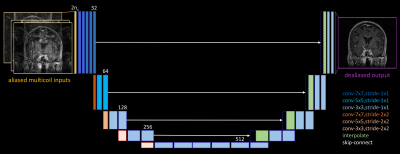
Network design. We studied neural networks that operate entirely in the image domain, taking aliased multicoil images as input and producing a coil-combined dealiased image as output. We reduced the 3D reconstruction problem into a sequence of 2D problems by separating along the fully-sampled readout axis. This simplified training and produced more training points.
Network training. We set aside 3 patient datasets of varying pathological rarity for testing, and trained using the other 45 datasets. We separated along the readout axis aliased coil images and corresponding root-sum-of-squares fully-sampled reconstructions, yielding $$$\sim$$$11,000 2D input-target pairs. Using a standard 90%/10% update/validation training data split to avoid overfitting, we trained more than 40 different model-free networks, varying topologies (encoder-decoder, convolutional, u-net, etc.), depths (4-40+ layers), patch sizes (8x8 to full-FOV), and sampling patterns (regular, variable-density). When run to convergence, training instances each required 3-5 days of wall time on a Tesla P40 GPU with 24GB RAM. We report results on a 35-layer u-net4 (with full-FOV patches and regular 2x2 undersampling) that achieved similar test performance across several training instances.
Results
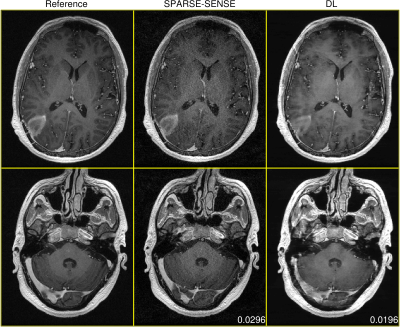
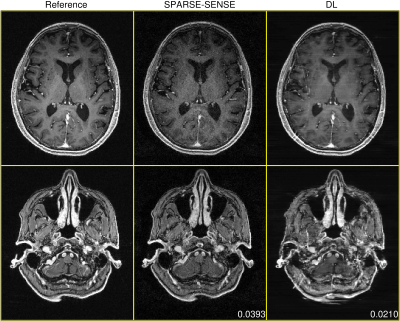
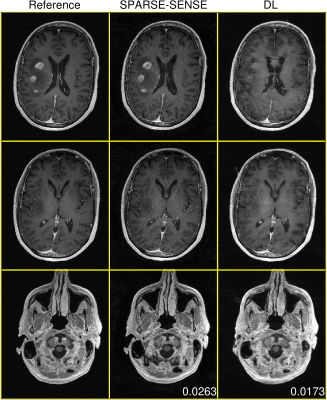
Figures 2-4 demonstrate DL test performance on 3 cases of increasing pathological rarity. Results are presented alongside a root-sum-of-squares reconstruction from fully-sampled data for reference, as well as a joint compressed sensing and SENSE reconstruction (SPARSE-SENSE5) at the same undersampling level for comparison. All full-volume DL reconstructions required less than 10 seconds on the aforementioned hardware. In easier cases (e.g., Figures 2-3), DL is able to reconstruct reasonably, though residual aliasing and blurring is sometimes apparent. However, in rare pathologies unseen in training (e.g., Figure 4), DL can make strong errors that may distort or even hallucinate away key anomalies. Interestingly, DL still achieves lower mean absolute error (MAE) than SPARSE-SENSE in all cases.Conclusion
This work has investigated the feasibility and robustness of model-free DL reconstruction. We have observed that while model-free DL can reasonably reconstruct anatomies similar to those seen in training, performance can degrade drastically for more challenging cases.Acknowledgements
No acknowledgement found.References
[1] Zhang et al., arXiv 1906.10643v1, 2019.
[2] Hammernik et al., MRM, 2017.
[3] Zhang et al., MRM, 2013.
[4] Ronneberger et al., Proc. MICCAI, 2015.
[5] Otazo et al., MRM, 2010.
Figures