3577
Fetal Brain Motion Estimation to Evaluate Motion-induced Artifacts in HASTE MRI1Department of Engineering Physics, Tsinghua University, Beijing, China, 2Department of Electrical Engineering and Computer Science, Massachusetts Institute of Technology, Cambridge, MA, United States, 3Computer Science and Artificial Intelligence Laboratory (CSAIL), Massachusetts Institute of Technology, Cambridge, MA, United States, 4Fetal-Neonatal Neuroimaging and Developmental Science Center, Boston Children’s Hospital, Boston, MA, United States, 5Harvard Medical School, Boston, MA, United States, 6Institute for Medical Engineering and Science, Massachusetts Institute of Technology, Cambridge, MA, United States
Synopsis
Artifacts generated by severe and unpredictable fetal and maternal movements during MRI limit the success of imaging during pregnancy. Although modern clinical applications use single-shot imaging sequences, such as HASTE, to partially mitigate this problem, inter-slice motion artifacts are unavoidable and their impact on fetal imaging is not fully characterized. In order to analyze this problem, we exploit a large repository of volumetric EPI over long duration across many pregnant women to estimate the artifact load due to inter-slice motion on single-shot fetal brain MRI.
Introduction
Fetal motion is the most challenging source of artifacts in fetal MR imaging. In order to reduce the impact of fetal motion, Half Fourier Acquisition Single Shot Turbo Spin Echo (HASTE) is often used in an attempt to “freeze” fetal motion. Although the scanning time of a single HASTE slice is short (1.6-1.8s), SAR constraints require a substantial time interval delay between slices (1.1-1.3s), thus increasing the overall scan time for a multi-slice volume and increases the likelihood of inter-slice motion-derived imaging artifacts. To investigate and characterize the severity of inter-slice motion artifacts on fetal brain imaging, we performed a simulated scan based on long-duration (10-30min), volumetric EPI observations of pregnant mothers. The simulation captures motion patterns that lead to fetal brain imaging artifacts in single-shot HASTE, and allows us to quantify the motion induced missed volumes and spin history artifacts on fetal brain HASTE imaging.Methods
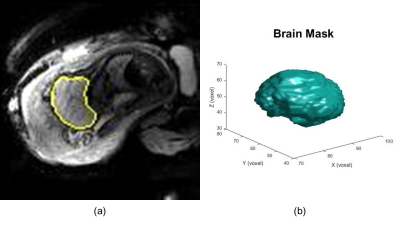
Our volumetric data repository contains multislice EPI volumes1 (matrix size = 120*120*80, resolution = 3mm*3mm*3mm, time sampling rate = 2.0-4.0s, imaging time between 10 min and 30 min) of the pregnant abdomen for fetuses with gestational age between 25 and 35 weeks. A 3D U-Net2 was applied to segment the fetal brain and obtain the volume of the fetal brain in each TR. From a total of 111 subjects, after excluding twin pregnancies, 24 subjects were selected based on the accuracy of fetal brain segmentation3 (segmented brain volume variance is 3%-10%). These relatively low spatial resolution MR volumes have adequate image fidelity to capture fetal body and brain motion over time, and the EPI TR is close to that used in single-shot HASTE. Under a quasi-static assumption, the fetus was assumed to be moving from one HASTE TR to the next, thus simulating the effect of inter-slice artifacts.A plurality of EPI images in a fixed length of time were considered as a group where the first segmented brain volume was considered a reference volume. The motion information included in subsequent frames of the time series was utilized for simulated single-shot imaging, and thus the analysis was performed for the difference between the orientation of subsequent brain volumes (simulated scanning slices) and the reference brain volume (simulated initial slice).
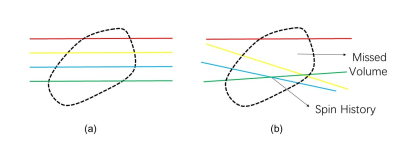
Fetal brain motion information included in the differences between consecutive images in time series was based on ITK4 to extract motion information. The images were registered5 based on the brain mask obtained by brain segmentation to derive the rigid-body transformation information of the fetal brain over time. Simulated single-shot imaging with standard slice thickness and TR was prescribed in the scanner coordinate system at the beginning of each group of frames and the proportion of missed volume (gaps between slices) and spin history (overlapping slices) in the reference volume were used to quantify inter-slice motion artifacts.
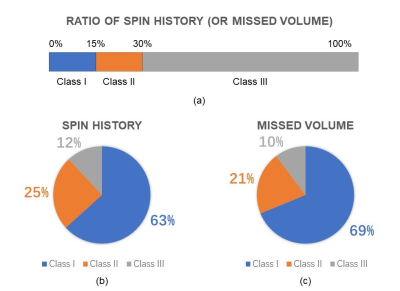
We analyzed more than 500 sets of fetal brain motion models for a simulated total scan time of 8 hours of single-shot fetal brain MRI. Through the missed volume and spin history metrics, we rated the simulated scan quality of each model. We defined the data whose missed volume and spin history were larger than 30% as class III; both metrics below 15% as class I; and others as class II.
Results and Discussion
The results in Figure 3 shows that by these metrics and thresholds, approximately 1/3 of the data was estimated to be of questionable quality (class II and class III).Although the missed volume and spin history ratio as the evaluation criteria are affected by image resolution, the segmentation accuracy of our U-Net networks trained with over 500 data models suggests that improving resolution will have little impact on the overall conclusion. In the fetal brain coordinate system, missed volume and spin history result from the variation of scanning position and angle due to fetal motion. The imaging quality of the simulated scan and the degree that the inter-slice artifacts matter can be evaluated by these metrics. However these metrics capture only two contributing factors to poor HASTE image quality and does not capture intra-slice motion artifacts, shimming errors, or RF-related artifacts that may be amplified by maternal or fetal motion and adversely affect diagnostic quality of fetal MRI. In addition, this approach does not account for double oblique acquisitions which make clinical interpretation challenging.
Conclusion
We proposed a method to quantify the artifact load in commonly applied diagnostic single-shot T2-weighted fetal brain imaging due to missed volumes and spin history effects using motion transformations derived from low-resolution EPI volumes acquired over long scan durations (10-30 minutes) in pregnant subjects. The missed volume and spin history metrics offer a simple quantification of two types of motion-induced artifacts, but are an underestimate of the artifact load in fetal MRI. Future studies will include additional factors that impact fetal HASTE image quality discussed above.In addition, future work based on this research will explore how to design real time adjustment to slice prescription by tracking the brain based on pose estimation to mitigate the problem of inter-slice artifacts caused by fetal motion. This motion model could be used to guide the design of online, prospective motion detection and mitigation methods.
Acknowledgements
NIH R01 EB017337, U01 HD087211 and R01HD100009. NVIDIA Corporation.References
[1] Luo, J., Turk, E. A., Bibbo, C., Gagoski, B., Roberts, D. J., Vangel, M., ... & Barth, W. H. (2017). In vivo quantification of placental insufficiency by BOLD MRI: a human study. Scientific reports, 7(1), 3713.
[2] Ronneberger, O., Fischer, P., & Brox, T. (2015, October). U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical image computing and computer-assisted intervention (pp. 234-241). Springer, Cham.
[3] Hesamian, M. H., Jia, W., He, X., & Kennedy, P. (2019). Deep Learning Techniques for Medical Image Segmentation: Achievements and Challenges. Journal of digital imaging, 1-15.
[4] Yoo, T. S., Ackerman, M. J., Lorensen, W. E., Schroeder, W., Chalana, V., Aylward, S., ... & Whitaker, R. (2002). Engineering and algorithm design for an image processing API: a technical report on ITK-the insight toolkit. Studies in health technology and informatics, 586-592.
[5] Liao, R., Turk, E. A., Zhang, M., Luo, J., Grant, P. E., & Adalsteinsson, E., et al. (2016). Temporal registration in in-utero volumetric mri time series. Med Image Comput Comput Assist Interv, 54-62.
Figures