3563
Fully automatic three dimensional prostate images registration: from T2-weighted-images to diffusion‐weighted images1Peking University, Beijing, China, 2Peking University First Hospital, Beijing, China
Synopsis
In the wake of population aging, prostate cancer has become one of the most important diseases in western elderly men. T2-weighted-images (T2WIs) provides the best depiction of the prostate’s anatomy, and diffusion‐weighted images (DWIs) provides clues of cancer as prostate cancer appears as an area of high signal on DWIs. However, high b-value DWIs is affected by the susceptibility effects. In this paper,we describe and utilize a fully automatic 3D deep neutral network to correct misalignment between T2WIs and DWIs. The average increasement of Dice coefficient of before-and-after registration is 73% and 84%, which reflects the removal of susceptibility effects.
Introduction
In the wake of population aging, prostate cancer has become one of the most important diseases in western elderly men, especially in United State [1]. Multi-parametric MRI is the most common used non-intrusive technique to diagnosis prostate cancer [2]. T2-weighted-images (T2WIs) provides the best depiction of the prostate’s zonal anatomy and capsule, and diffusion‐weighted images (DWIs) provides clues of cancer as prostate cancer appears as an area of high signal on DWIs.However, high b-value DWIs is affected by geometric distortions and chemical shift artifacts caused by the susceptibility effects. These effects are due to the air-filled balloon surrounding the endorectal coil and poor local B0 homogeneity which leads to pixel shifts, particularly in the phase encode direction. For a fusion system to work effectively, accurate registration of different imaging modalities is critical. However, multi-modality image registration is a very challenging task, as it is hard to define a robust image similarity metric .
Recently, the widespread adoption of deep learning techniques has led to remarkable achievements in the field of multi-modality image registration. Many deep learning based approaches for registration have used convolution neural network (CNN) regressors to estimate deformation field [3,4]. In this paper, we introduce a fully automatic 3D network architecture to jointly perform deformable image registration.
Image Acquisitions
Clinical routine MRI was conducted on a cohort of 50 subjects, including healthy subjects, prostate cancer patients and prostate hyperplasia patients. The imaging was performed on a 3.0 T Ingenia system (Philips Healthcare, the Netherlands), with acquisition of T2-weighted imaging and DWI. T2w turbo-spin-echo images were obtained in the axial, sagittal, and coronal planes without fat suppression with TR: 2905 ms; TE: 90 ms; FOV: 240 mm×240 mm; matrix: 324×280; slice thickness: 4 mm with no gap. DWI were also obtained in the axial, sagittal, and coronal planes with TR: 3970 ms; TE: 68 ms; FOV: 240 mm×240 mm; matrix: 184×184; slice thickness: 4 mm with no gap; two b values (b0, b1000).Study Population
We performed this retrospective study with permission from the local Institutional Ethical Committee. The need for informed consent was waived. From these patients, the anatomical landmarks (full gland segmentations) were manual outlined by two experts with more than five years’ experience.The inhouse dataset was splitting into training dataset, validation dataset and testing dataset as 60%, 20% and 20% of all 50 subject, respectively. Before training, each dataset was transformed by a random affine transformation for data augmentation.
Algorithm description
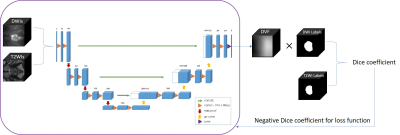
In this work, the entire training procedure involves for major steps.Firstly, a pair of multimodal 3D blocks (a T2WI 3D block and a DWI 3D block image) were input to the network without any segmentation labels, the network was designed as a classical 3D U-net that generates dense displacement vector field (DVF) from input images. The design of DVF was aimed to give descriptions of deformation strength and direction for each pixel.
Secondly, the learned DVF was applied to the DWI block to get the registered-DWI block.
Thirdly, to evaluate the anatomy difference between the registered-DWI block and the T2WI block, the manual segmentation label of the DWI block was deformated by same DVF. Then the registration accuracy was gained by computing Dice coefficient between the registered-DWI-label and T2WI-label.
Finally, the negative Dice coefficient was used as loss function of the network to increase performance of DVF.
For predicting procedure, only the first and second step was required.
Result
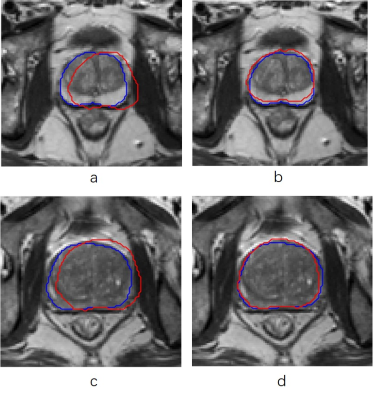
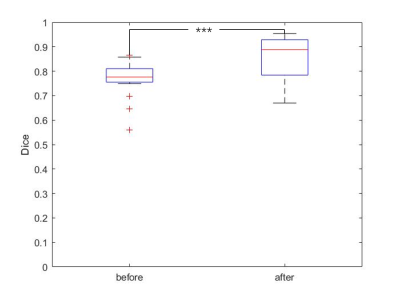
In Fig. 2 we shows some examples of before and after registration result in T2WI image.Better registration is reflected by closer alignment of the two contours.The registration performance of the developed method was then quantitatively evaluated and the results are given in Fig. 3. The evaluation was performed using Dice Score caculated by the T2 and DWI images, respectively. It can be seen that the Dice socre increase significantly after register(from 0.73 to 0.84 ,p <0.01)Discussion and Conclusion
In this paper,we describe an architecture utilizing a deep neutral network to correct misalignment between T2WIs and DWIs. The proposed method targets a wide range of clinical applications, where automatic multimodal image registration has been traditionally challenging due to the lack of reliable image similarity measures or automatic landmark extraction methods. Unlike conventional registration methods, our network encodes and learns the most relevant features for joint image registration. What's more, the anatomical labels are only used in training to evaluate images similarity measures, which would result in a much amenable in clinical practice. The future works could concentrate on multiple organ registration as more organ label may provide more anatomical information.Acknowledgements
No acknowledgement found.References
[1]Siegel R, Ma J, Zou Z, et al. Cancer statistics, 2014[J]. CA: a cancer journal for clinicians, 2014, 64(1): 9-29.
[2]Villers A, Lemaitre L, Haffner J, et al. Current status of MRI for the diagnosis, staging and prognosis of prostate cancer: implications for focal therapy and active surveillance[J]. Current opinion in urology, 2009, 19(3): 274-282.
[3]Elmahdy M S, Wolterink J M, Sokooti H, et al. Adversarial optimization for joint registration and segmentation in prostate CT radiotherapy[C]//International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Cham, 2019: 366-374.
[4]Hu Y, Modat M, Gibson E, et al. Weakly-supervised convolutional neural networks for multimodal image registration[J]. Medical image analysis, 2018, 49: 1-13.
Figures