3561
Whole Knee Cartilage Segmentation using Deep Convolutional Neural Networks for Quantitative 3D UTE Cones Magnetization Transfer Modeling
Yanping Xue1,2, Hyungseok Jang1, Zhenyu Cai1, Hoda Shirazian1, Mei Wu1, Michal Byra1, Yajun Ma1, Eric Y Chang1,3, and Jiang Du1
1University of California, San Diego, San Diego, CA, United States, 2Beijing Chao-Yang Hospital, Beijing, China, 3VA San Diego Healthcare System, San Diego, CA, United States
1University of California, San Diego, San Diego, CA, United States, 2Beijing Chao-Yang Hospital, Beijing, China, 3VA San Diego Healthcare System, San Diego, CA, United States
Synopsis
The existence of short T2 tissues and high ordered collagen fibers in cartilage render it “invisible” to conventional MR and sensitive to the magic angle effect. Segmentation is the first step to obtain parameters of cartilage, which is often performed manually (time-consuming and variable). Automatic segmentation and providing a biomarker that visualizes both short and long T2 tissues and insensitive to the magic angle effect is desideratum. U-Net is based on CNN to process images. The purpose of this study is to describe and evaluate the pipeline of fully-automatic segmentation of cartilage and extraction of MMF in 3D UTE-Cones-MT modeling.
Introduction
Osteoarthritis (OA) affects millions of people and has a substantial impact on the economy and the health care system worldwide. Various cartilage pathologies, such as the depletion of proteoglycan (PG) or degeneration of the collagen network, are directly associated with the onset and progression of knee OA [1]. Many quantitative MRI biomarkers have been used to detect the early degeneration of cartilage, but because of the existence of short T2 tissues and the highly ordered collagen fibers in cartilage which make conventional MR “invisible” and sensitive to magic angle effect and limit conventional MR parameters to detect the early biochemical changes in these regions [2-4]. Furthermore, in order to obtain quantitative MR measurements of cartilage in the knee joint, the first step is the segmentation of the cartilage, which is often performed manually and is therefore time-consuming and usually prone to the inter-observer variability and bias [5]. Therefore, there has been great interest in developing an accurate and fully automatic method to segment cartilage, which allows a seamless workflow to provide a quantitative biomarker visible both short and long T2 tissues and less sensitive to magic angle effect. With the recent innovation in computational power of CPU and GPU with decreased cost allowed the deep learning to be widely used in many applications [6-11]. U-Net is based on deep convolutional neural networks (CNN), known as effective networks to process images, which is based on a fully-convolutional network comprised of two main paths: encoder (or contracting) path and decoder (or expansive) path. Shortcut connections between the layers are commonly added to improve object localization [12]. The purpose of this study is to describe and evaluate the pipeline of fully automatic segmentation of whole knee cartilage and extraction of MMF with deep CNN in 3D UTE-Cones-MT modeling.Methods
A total of 65 human subjects (aged 20-88 years; 54.8±16.9 years; 32 males, 33 females) was recruited for this study. Informed consent was obtained from all subjects in accordance with the guidelines of the Institutional Review Board. The whole knee joint (29 left knees, 36 right knees) was scanned using 3D UTE-Cones-MT sequences (saturation pulse power = 500°, 1000°, 1500°; frequency offset = 2, 5, 10, 20, 50 kHz; flip angle = 7˚) on a 3T MR750 scanner (GE Healthcare Technologies, Milwaukee, WI). The architecture of the attention U-Net CNN for cartilage segmentation is presented in Fig.1. In comparison to the standard U-Net CNN, our network included attention gates that process feature maps propagated through the skip connections from the encoder path. MMF was calculated using a two-pool MT model. The Dice coefficient and the volumetric overlap error (VOE) were used to evaluate the accuracy of cartilage segmentation between labels from the manual and automatic segmentation.Results and Discussion
Fig. 2 shows the results with two representative subjects (A: 72-year-old female, B: 43-year-old male), displaying the input UTE-MR image and the corresponding manual labels by two radiologists (Rad1 and Rad2) and the resultant labels produced by CNN (CNN1 and CNN2). Strong inter-observer agreement was shown between Rad1 and CNN1 or between Rad2 and CNN2, where the morphology of the labels by each CNN showed high similarity with the labels by the corresponding radiologist. Table 1 shows the Dice coefficients of 0.84 and 0.77 for Rad1 vs CNN1 and Rad 2 vs CNN2, respectively. The Dice score between the labels produced by the CNN1 and CNN2 was 0.79, which was higher than the Dice score (0.71) between the two radiologists, indicating the improved inter-observer agreement in the automatic segmentation than the manual segmentation. The Dice score was 0.76 between the Rad 1 and CNN 2 and 0.72 between the Rad2 and CNN1, which was a little better than that between the radiologists. The VOE between the manual and automatic segmentations was 43.69%±13.15%, 27.13%±9.91%, 42.29% ±12.76%, 38.11%±10.10%, 36.68%±11.91%, and 33.82%±9.38% for Rad1vs.Rad2, Rad1 vs CNN1, Rad2 vs CNN1, Rad1 vs CNN2, Rad2 vs CNN2, and CNN1 vs CNN2, respectively. Table 2 lists the average values of MMF calculated based on the manual and automatic segmentation, respectively. The results of ICC of MMF estimated using the manual and automatic segmentations reached a high level of consistency: the ICC was equal to 0.94, 0.86, 0.80, and 0.92, for the Rad1vs.CNN1, Rad2vs.CNN2, Rad1 vs Rad2, and CNN1vs.CNN2, respectively. Fig. 3 shows the scatter plots for MMF values between the manual and automatic segmentation. The Pearson’s linear correlation coefficients for MMF values depicted in Figure 3a-d between the Rad1vs.CNN1, Rad2vs.CNN2, Rad1vs.Rad2, and CNN1vs.CNN2 were equal to 0.89, 0.78, 0.68, and 0.87, respectively. All obtained correlation coefficients were high (p-values<0.001), indicating a high level of agreement between the radiologists and the deep learning models.Conclusion
The proposed transfer learning-based U-Net CNN model can automatically segment whole knee cartilage and extract the quantitative MT parameters in 3D UTE-Cones MR imaging. To the best of our knowledge, this is the first study to segment the whole knee cartilage using deep CNN with transfer learning in 3D UTE-Cones-MT modeling for accurate assessment of the MMF in cartilage.Acknowledgements
The authors acknowledge grant support from NIH (R01AR075825, 2R01AR062581, 1R01 AR068987), and the VA Clinical Science and Rehabilitation R&DAwards (I01RX002604)References
1. Hani AF, Kumar D, Malik AS, et al. Non-invasive and in vivo assessment of osteoarthritic articular cartilage: a review on MRI investigations. Rheumatol Int. 2015, 35(1):1-16. 2. Du J, Carl M, Bae WC, et al. Dual inversion recovery ultrashort echo time (DIR-UTE) imaging and quantification of the zone of calcified cartilage (ZCC). Osteoarthritis Cartilage. 2013; 21(1):77–85. 3. Ma YJ, Shao H, Du J, Chang EY. Ultrashort echo time magnetization transfer (UTE-MT) imaging and modeling: magic angle independent biomarkers of tissue properties. NMR Biomed 2016, 29(11):1546–1552. 4. Ma YJ, Carl M, Searleman A, et al. 3D adiabatic T1ρ prepared ultrashort echo time cones sequence for whole knee imaging. Magn Reson Med. 2018, 80(4):1429-1439. 5. Liu F, Zhou Z, Jang H, et al. Deep convolutional neural network and 3D deformable approach for tissue segmentation in musculoskeletal magnetic resonance imaging. Magn Reson Med. 2018, 79(4):2379-2391. 6. Raina R, Madhavan A, Ng AY. Large-scale deep unsupervised learning using graphics processors. In: Proceedings of the 26th Annual International Conference on Machine Learning - ICML ’09. Montreal, Canada, 2009, pp. 1-8. 7. Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. In: The IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2015, pp: 3431-3440. 8. Hinton G, Deng L, Yu D, et al. Deep neural networks for acoustic modeling in speech recognition: the shared views of four research groups. IEEE Signal Processing Magazine. 2012, 29:82-97. 9. Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. ArXiv: 1409.1556, 2014. 10. Badrinarayanan V, Kendall A, Cipolla R. SegNet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans Pattern Anal Mach Intell. 2017, 39(12):2481-2495. 11. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015, 521(7553):436–444. 12. Byra M, Wu M, Zhang X, et al. Knee menisci segmentation and relaxometry of 3D ultrashort echo time cones MR imaging using attention U-Net with transfer learning. Magn Reson Med. 2019, DOI: 10.1002/mrm.27969.Figures
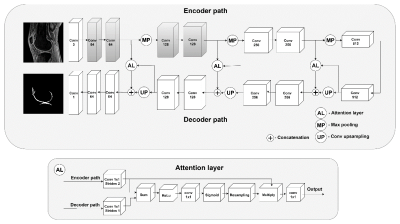
Fig. 1 The
proposed 2D attention U-Net CNN for the cartilage segmentation. Note that the
blocks in gray color indicate the weights initialized from the VGG19 network
for transfer learning.

Fig. 2 The T1ρ MR images obtained using the 3D UTE cones for
NIR=2 and the whole knee cartilage segmented using the manuals and CNN models,
respectively. Row A: (72-year-old female), the medial side of the knee and
segmented cartilage; Row B: (43-year-old male), the lateral side of the knee
and segmented cartilage. Solid arrows indicate the difference between the CNN1
and CNN2, while dashed arrows indicate the difference between Rad1 and Rad2.

Table 1 The Dice coefficients and the volumetric overlap error (VOE)
(mean ± SD) calculated using the manual and automatic segmentation.

Table 2 The values of MMF (mean ± SD) calculated using the manual and
automatic segmentations, and the consistency evaluation with ICC between the
radiologists and CNN models.
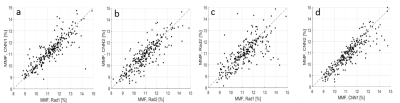
Figure 3 Scatter
plots for the MMF values between the manual and automatic segmentation. The
Pearson’s correlation coefficients for MMF values depicted in between the Rad 1
vs CNN 1 (a), Rad 2 vs CNN 2 (b), Rad 1 vs Rad 2 (c), and CNN 1 vs CNN 2 (d) are
equal to 0.89, 0.78, 0.68, and 0.87, respectively. All obtained correlation
coefficients are high (p-values<0.001), indicating a high level of
agreement between the radiologists and the deep learning models.