3559
Automated Segmentation of Late Gadolinium Enhanced CMR with Deep Learning1Biomedical Engineering, Northwestern University, Evanston, IL, United States, 2Radiology, Northwestern University Feinberg School of Medicine, Chicago, IL, United States, 3Feinberg Cardiovascular Research Institute, Northwestern University Feinberg School of Medicine, Chicago, IL, United States
Synopsis
Late gadolinium enhanced (LGE) CMR is the gold standard test for assessment of myocardial scarring. While quantifying scar volume is helpful to clinical decision making, its lengthy image segmentation time limits its use in practice. The purpose of this study is to enable fully automated LGE image segmentation using deep learning (DL) and explore a more efficient way of using annotation.
Introduction
Late Gadolinium Enhancement (LGE) cardiovascular magnetic resonance (CMR) is the gold standard test for assessment of myocardial scarring (1). While scar volume quantification is potentially a useful predictor of major adverse cardiac event (MACE), it is rarely used in clinical practice due to lengthy image segmentation time. In this study, we sought to develop a fully automated approach for LV scar quantification using deep learning (DL) in a more efficient way of utilizing the annotated data.Methods
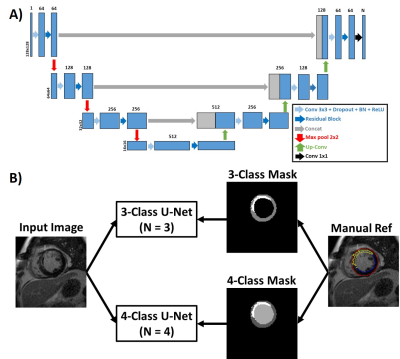
Human Subject: This study leveraged our access to labeled LGE data with myocardial wall and scar contours from the DETERMINE Trial (ClinicalTrials.gov ID NCT00487279). In total, 101 patients with a history of CAD and mild to moderate left ventricular dysfunction from 5 different hospitals were included in this study.Image Processing: As shown in Figure 1, we implemented a multi-class residual U-Net (2) on a GPU workstation (NVIDIA Tesla V100) equipped with Pytorch, in order to automatically segment the whole left ventricle into 4 classes: background, healthy myocardium, scar, and LV blood pool. Labeled data were randomly selected from 81 patients (764 2D images in total) were used for training the network, and the remaining labeled data from 20 patients (184 2D images in total) were used for testing. Manual contour files included endo-myocardium, epi-myocardium border and scar information were processed in MATLAB and converted into masks with labels on each pixel (i.e. background = 0; healthy myocardium = 1; LV blood pool = 2; scar = 3). The magnitude images and the labeled masks were used as input/output pairs. All images and masks were cropped to 128x128 matrix size to keep consistent image size. We used Pytorch built-in cross entropy loss combined with dice loss of the whole LV (i.e. Myocardium outer boundary). Optimization was performed using Adam optimizer with learning rate of 0.001 and 100 epochs. We compared our approach to a previously described method (3) using 3 classes (background = 0; healthy myocardium = 1; scar = 2), where both our approach and previous approach used the same manual contour files, as shown in Figure 2. The extra labeling of blood pool cost no additional editing of the contours.
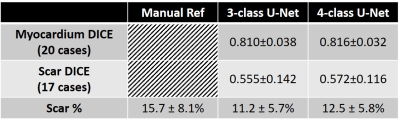
Quantitative Analysis: To evaluate the network performance, we calculated Sørensen–Dice index and scar volume (percentage of scar tissue over myocardium) for each testing subject with manual contours as reference. For myocardium DICE score, all 20 testing cases were included, while for scar DICE score and scar volume, 3 cases that doesn’t have scar were excluded.
Results
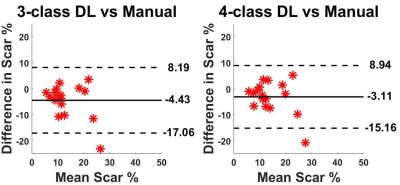
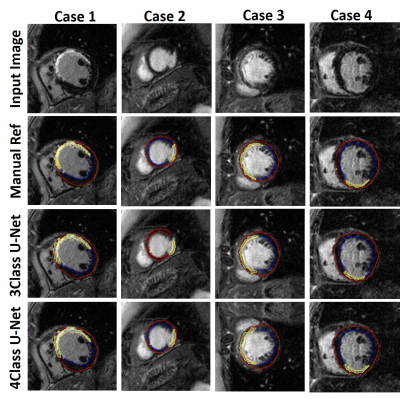
Figure 3 shows results from 4 representative patients, in which 4-class network was slightly better than 3-class network with better connectivity of segmented regions. As summarized in Table 1, 4-class network produced slightly higher mean DICE scores for myocardium and scar than 3-class network, but they were not significantly different (P>0.05). The scar volume was not significantly (P>0.05) different between manual, 3-class U-Net, and 4-class U-net using 1-way ANOVA.Discussions
Our proposed 4-class U-Net showed the potential of automatic segmentation of the whole LV for LGE images, which showed better results than 3-class U-Net model. The extra labeling of LV blood pool as an individual class instead of background improved the performance of deep learning network. Even though the blood pool was not our target for segmentation, labeling it could help the targeted segmentation of scar. Future study is warranted to further explore the best way of utilizing the annotations and get better segmentation results.Acknowledgements
This work was supported in part by the following grants: National Institutes of Health (R01HL116895, R01HL138578, R21EB024315, R21AG055954) and American Heart Association (19IPLOI34760317).References
1. Simonetti OP, Kim RJ, Fieno DS, Hillenbrand HB, Wu E, Bundy JM, Finn JP, Judd RM. An improved MR imaging technique for the visualization of myocardial infarction. Radiology 2001;218(1):215-223.
2. Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. 2015. Springer. p 234-241.
3. Fahmy AS, Rausch J, Neisius U, Chan RH, Maron MS, Appelbaum E, Menze B, Nezafat R. Automated cardiac MR scar quantification in hypertrophic cardiomyopathy using deep convolutional neural networks. JACC: Cardiovascular Imaging 2018;11(12):1917-1918.
Figures