3556
Unsupervised learning for Abdominal MRI Segmentation using 3D Attention W-Net1Faculty of Computer Science, Otto von Guericke University, Magdeburg, Germany, 2Department of Biomedical Magnetic Resonance, Otto von Guericke University, Magdeburg, Germany, 3Data & Knowledge Engineering Group, Otto von Guericke University, Magdeburg, Germany, 4Center for Behavioral Sciences, Magdeburg, Germany, 5German Center for Neurodegenerative Disease, Magdeburg, Germany, 6Leibniz Insitute for Neurobiology, Magdeburg, Germany
Synopsis
Image segmentation is a process of dividing an image into multiple coherent regions. Segmentation of biomedical images can assist diagnosis and decision making. Manual segmentation is time consuming and requires expert knowledge. One solution is to segment medical images by using deep neural networks, but traditional supervised approaches need a large amount of manually segmented training data. A possible solution for the above issues is unsupervised medical image segmentation using deep neural networks, which our work tries to solve with our proposed 3D Attention W-Net.
Introduction
Image Segmentation is the process of dividing the image into multiple coherent regions, where the pixels in each segment are contextually connected. Many approaches to medical image segmentation have been proposed. These methods require a large amount of truthfully segmented training data [1][2]. Abdominal MR image segmentation is an interesting research area, and not yet very much explored, especially when less to no ground truth is available. Our state of the art model is based on a W-Net [3] with both U-Nets replaced by attention U-Nets [4], and also the model has been extended to work with 3D volumes.Method
In this research, the input image intensities are normalized first before giving it to the network to bring it to a common scale for faster convergence while training. Simultaneously, the weights between the voxels are calculated using Eq-1, where wij is the weight between the pixel i and j, which is required in calculating Normalized-Cuts using Eq-2 (loss function).$$w_{i\ j}=e^{\frac{-\|F(i)-F(j)\|_{2}^{2}}{\sigma_{I}^{2}}}\ \ast\left\{\begin{array}{cc}{\frac{-\|X(i)-X(j)\|_{2}^{2}}{\sigma_{X}^{2}}}\ &\ \end{array}\right.(Eq.1)[3]
$$\begin{aligned} J_{\text {soft-Ncut}}(V, K) &= K-\sum_{k=1}^{K} \frac{\sum_{u \in V} p\left(u=A_{k}\right) \sum_{u \in V} w(u, v) p\left(v=A_{k}\right)}{\sum_{u \in V} p\left(u=A_{k}\right) \sum_{t \in V} w(u, t)} (Eq.2)[3] \end{aligned}
The architecture is based on auto-encoders, in which the encoder part maps the input to the pixel-wise segmentation layer without losing its original spatial size despite low-dimensional space and the decoder part reconstructs the original input image from the dense prediction layer.
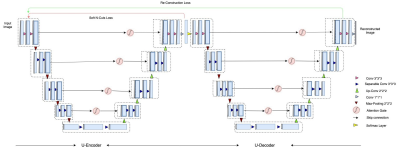
Model Construction - 3D Attention W-Net
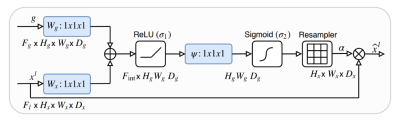
The base W-Net architecture proposed by [3] is modified by replacing both U-Nets with recently proposed attention U-Nets [4] (Attention gate architecture shown in Figure 2). Both, W-Net and attention U-Net have been proposed for 2D images. In our case, we have modified the network architecture to work with 3D volumes. Although 3D image segmentation can benefit many use cases, there has not been much progress in the context of 3D MR images, until recently, but all of the approaches are based on supervised learning. Our state of the art model works with 3D images, such as MR volumetric data. Our network consists of 18 modules in which each module consists of two 3D convolutional layers with kernel size 3x3x3. Each layer is followed by a non-linear activation function (PReLU) and an instance normalization layer. In total, we are using 46 3D convolutional layers. The first 9 modules represent the encoder network in which the network predicts the segmentation maps and the next 9 modules reconstruct the original input image from the low dimensional output of the encoder part. In the contraction path, the modules are connected through max pool 3D layers and in the expansion path, modules are connected through upsample layers followed by convolutional layers. Skip connections are passed through attention gates to suppress irrelevant regions and noisy responses. The output of an encoder is given to 3D convolution layers with kernel size 1x1x1, stride=1 and filter = 1 followed by a softmax layer. The second U-Net is similar to the first one, with the only difference being the final activation function. A sigmoid function has been used instead of softmax. We trained our network with the MR abdominal dataset provided by [5], which contains 40 volumes including manually segmented ground-truth. The given set was divided into training (25 vols), validation (5 vols) and test sets (10 vols). We train our model such that it minimizes both N-cuts (loss for only the first U-Net) and reconstruction loss (loss for both U-Nets). Since we use many CNN's with max-pooling layers in the model, this results in increased invariance, which can cause localization accuracy reduction. To prevent the fine boundaries in the output segments, we used CRF [9] as a post-processing step. Our proposed network architecture has been shown in Figure 1.Results
We used both U-Nets for training the network on a given training dataset and while testing the given image set, we used only first U-Net. The output of the first U-Net is the required segmentation output. So, we neglect the second u-net while validating. The predicted segmentation is then passed through CRF (Conditional Random Fields) post-processing technique to recover the boundaries. Since there is no other work which is to compare our results with, we compare our results with the available ground-truth. We considered only the liver as our region of interest, so we have taken only the clusters that provide the liver segment into consideration from both output and ground-truth. One example result and quantitive analysis in terms of Intersection over Union and Dice Coefficient are shown in Figures 3 and 4 respectively.Conclusion
In this work, for the problem of medical image segmentation, we have proposed a model which segments abdominal 3D MRI volumes without any manual labeling information. We trained and tested our model with T1 weighted in-phase images. We extend the research of unsupervised segmentation of ordinary images using W-Net for segmentation of abdominal MRIs. We further enhanced our model by using Attention U-Nets and constructed a novel 3D Attention W-Net. It was observed that the model performs well. Ground truth images supplied in the dataset seems to include large liver vessels, but the network correctly segmented the liver and didn’t include them.Acknowledgements
This work was in part conducted within the context of the International Graduate School MEMoRIAL at OvGU (project no. ZS/2016/08/80646).References
[1] Vijay Badrinarayanan, Alex Kendall, and Roberto Cipolla. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE transactions on pattern analysis and machine intelligence, 39(12):2481–2495, 2017.
[2] Adam Paszke, Abhishek Chaurasia, Sangpil Kim, and Eugenio Culurciello. Enet: A deep neural network architecture for real-time semantic segmentation. arXiv preprint arXiv:1606.02147, 2016.
[3] Xide Xia and Brian Kulis. W-net: A deep model for fully unsupervised image segmentation. arXiv preprint arXiv:1711.08506, 2017.
[4] Ozan Oktay, Jo Schlemper, Loic Le Folgoc, Matthew Lee, Mattias Heinrich, Kazunari Misawa, Kensaku Mori, Steven McDonagh, Nils Y Hammerla, Bernhard Kainz, et al. Attention u-net: Learning where to look for the pancreas. arXiv preprint arXiv:1804.03999, 2018.
[5] “Chaos grand challenge.” website. Available online at https://chaos.grand-chalenge.org/Data/ visited on May 20th, 2019.
Figures