3555
Attention-based Semantic Segmentation of Thigh Muscle with T1-weighted Magnetic Resonance Imaging1School of Computer Science, University of Sydney, Sydney, Australia, 2Sydney Neuroimaging Analysis Centre, Sydney, Australia, 3Brain and Mind Centre, University of Sydney, Sydney, Australia, 4Discipline of Exercise and Sport Science, Faculty of Medicine and Health, University of Sydney, Sydney, Australia
Synopsis
Robust and accurate MRI-based thigh muscle segmentation is critical for the study of longitudinal muscle volume change. However, the performance of traditional approaches is limited by morphological variance and often fails to exclude intramuscular fat. We propose a novel end-to-end semantic segmentation framework to automatically generate muscle masks that exclude intramuscular fat using longitudinal T1-weighted MRI scans. The architecture of the proposed U-shaped network follows the encoder-decoder network design with integrated residual blocks and attention gates to enhance performance. The proposed approach achieves a performance comparable with human imaging experts.
Introduction
Neurological diseases such as multiple sclerosis may impact muscle strength and bulk. Interventions such as Neuromuscular Electrical Stimulation (NMES) cycling are designed to improve muscle strength1 and reduce disability. Precise thigh muscle segmentation with the exclusion of intra-muscular fat is critical to the quantification of longitudinal muscle volume change, a biomarker of treatment effect, in such studies.Traditional segmentation approaches are based on the intensity profile of the image and require human post-processing correct for image inhomogeneity and manually adjust the output (muscle) mask. To tackle this dilemma, we propose an end-to-end semantic deep learning framework for automated cross-sectional muscle segmentation from MRI scans. The proposed network learns residual information and deterministic contextual information in muscle regions. Evaluation of our method demonstrates muscle segmentation comparable with human expert labelled masks. Compared with manual labelling, which takes a human expert ~5 minutes to extract one cross-sectional muscle mask, the trained model automatically generates the predicted muscle mask in only ~0.1 seconds.
Methods
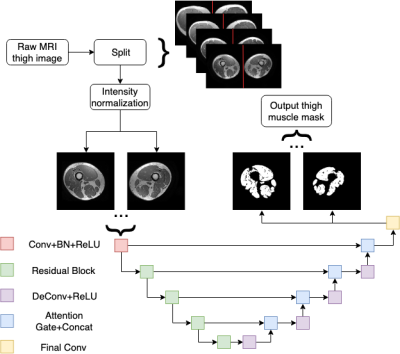
Eleven participants with progressive MS were recruited from a multidisciplinary MS clinic. MRI scans of both thighs of all participants were acquired on a GE discovery MR750 Scanner using a 32-channel torso coil with a 3DT1 sequence (IRFSPGR, TE = 2.7 msec, TR = 6.5 msec, acquisition matrix = 480 × 480, slice thickness = 1mm). The data collected includes scans from 31 time points in 11 participants.Following pre-processing steps are applied as described previously.2 Briefly, 1. Splitting the left and right thighs: each 3D MRI thigh image is firstly projected onto the axial plane by taking the maximum voxel values along the superior-inferior direction and dividing into left and right thigh respectively by locating the interpeak nadir in the associated intensity profile. 2. Subject-wise longitudinal registration: to obtain consistent FOV for muscle segmentation within subject, all followup scans were co-registered to baseline. Since the femurs are expected to be more morphologically stable than muscle over time, the registration transformation metrics were calculated based on femurs using FLIRT (FMRIB Software Library; www.fmrib.ox.ac.uk/fsl). 3. Intensity correction: intensity inhomogeneity-correction by N4 (Advanced Normalization Tools, http://picsl.upenn.edu/software/ants) and intensity normalization are applied to transform all data collected into a feasible format for subsequent training.
In summary, a total of 1290 cross-sectional thigh slices and corresponding manually labelled muscle masks were available for network training and validation purposes. The gold standard labelling was generated as previously described with manual QA.2 The pre-processed data was randomly split into a configuration of 72%, 9%, and 19% for training, validation, and testing, respectively.
The proposed model is based on the U-shaped encoder-decoder network design3 with skipped connections to share low-level information between the input and output (Figure 1). Instead of the standard convolutional strategy in the encoding procedure, we extend the convolution layers in the encoding path to residual blocks to enhance the learned information.4 Furthermore, attention gates5 are applied between the skipped connections to study contextual information in muscle regions. Although the traditional cross-entropy loss was used during training, we also employed the Dice loss which is more suitable for semantic segmentation tasks. The coding in this work is written in Python 3.6.9 (Python Software Foundation; https://www.python.org/) and PyTorch 1.1 (Facebook AI Research; https://pytorch.org/).
Results
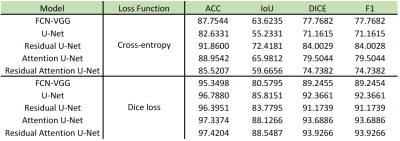
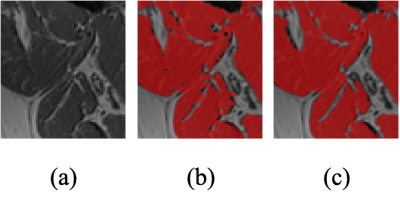
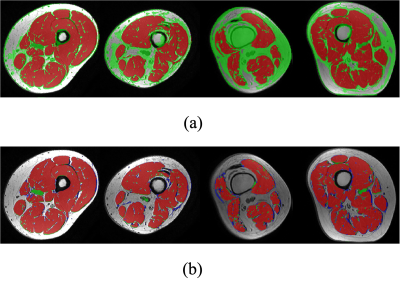
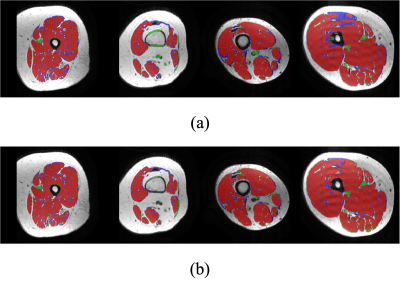
To evaluate the semantically segmented thigh muscle masks quantitatively, we devised an evaluation metric based on the mean value of pixel-wise accuracy (ACC), intersection over union (IoU), dice coefficient score (Dice), F1-score (F1). FCN-VGG6 is included as the benchmark for inference. The evaluation metrics facilitate the comparison of the proposed method with the labelled ground truth (Table 1).The ability of the trained network to exclude intra-muscular fat was qualitatively visualised (Figure 2). The muscle masks generated by different model designs (Figure 3) and loss functions (Figure 4), compared to the corresponding ground truth, were also visually evaluated for qualitative analysis. Visualization results include the pixels in both the predicted mask and the ground truth (true positives, marked in red), the pixels in the predicted mask but not in the ground truth (false positives, marked in green), and the pixels in the ground truth but not in the predicted mask (false negatives, marked in blue).
Discussion
Using Dice loss during the training phase more robustly distinguishes between foreground and background pixels by reducing a large number of false positives generated by employing cross-entropy loss. The effect of residual blocks and attention gates is shown both statistically and visually. Although the combination of these two modules does not improve performance when using cross-entropy as loss function, it ranks first among all components of the evaluation metric for Dice loss. Both quantitative and qualitative analyses demonstrated that the proposed framework reliably extracts muscle masks from T1-weighted MRI scans.Conclusion
In this work, we propose a novel end-to-end semantic segmentation framework to produce an accurate muscle mask from T1-weighted thigh MRI scans. The performance of our framework is comparable with human labelled results in terms of pixel-wise accuracy, IoU, Dice score, and F1-score. Our model is also capable of excluding intra-muscular fat and significantly improves time efficiency compared with manual workflows.Acknowledgements
No acknowledgement found.References
1. Fornusek C, Hoang P. Neuromuscular electrical stimulation cycling exercise for persons with advanced multiple sclerosis. Journal of rehabilitation medicine, 2014, 46(7): 698-702.
2. Tang Z, Wang C, Hoang P, et al. Automatic Segmentation of Thigh Muscle in Longitudinal 3D T1-Weighted Magnetic Resonance (MR) Images. Data Driven Treatment Response Assessment and Preterm, Perinatal, and Paediatric Image Analysis. Springer, Cham, 2018: 14-21.
3. Ronneberger O, Fischer P, Brox T. U-Net: Convolutional networks for biomedical image segmentation. International Conference on Medical image computing and computer-assisted intervention. Springer, Cham, 2015: 234-241.
4. He K, Zhang X, Ren S, et al. Deep residual learning for image recognition. Proceedings of the IEEE conference on computer vision and pattern recognition. 2016: 770-778.
5. Vaswani A, Shazeer N, Parmar N, et al. Attention is all you need. Advances in neural information processing systems. 2017: 5998-6008.
6. Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. CoRR, vol. abs/1409.1556, 2014.
Figures