3552
Adversarial Inpainting of Arbitrary shapes in Brain MRI1University of Stuttgart, Stuttgart, Germany, 2University Hospital Tübingen, Tübingen, Germany
Synopsis
MRI suffers from incomplete information and localized deformations due to a manifold of factors. For example, metallic hip and knee replacements yield local deformities in the resultant scans. Other factors include, selective reconstruction of data and limited fields of views. In this work, we propose a new deep generative framework, referred to as IPA-MedGAN, for the inpainting of missing or complete information in brain MR. This framework aims to enhance the performance of further post-processing tasks, such as PET-MRI attenuation correction, segmentation or classification. Quantitative and qualitative comparisons were performed to illustrate the performance of the proposed framework.
Introduction
Magnetic Resonance Imaging (MRI) is an essential component in modern medical procedures. However, MRI suffers from different localized deformities as a result of different factors. For instance, limited fields of views, superposition of foreign bodies (e.g. metallic implants1) in projection methods or the selective reconstruction of data are factors leading to localized deformations and missing information within MR scans. These artifacts render the MRI no longer suitable for diagnostic purposes. Despite this, automated image analysis algorithms would benefit from completing the missing or distorted regions2. One example is utilizing auto-completed MR for the calculation of attenuation coefficients for PET-CT attenuation correction. Similarly, auto-completion of MR scans is advantageous for more accurate dose calculation in radiation therapy planning.In this work, we propose a new deep learning framework for the inpainting of arbitrary-shaped missing or deformed regions in brain MRI scans. This should positively impact further post-processing tasks that emphasize global image properties rather than the detailed local or diagnostic information. The introduced framework, referred to as IPA-MedGAN, is based on previous Generative Adversarial Networks (GANs3-5) which consist of training multiple competing networks simultaneously in a game-theoretical approach aiming to reach Nash equilibrium.
Material and Methods
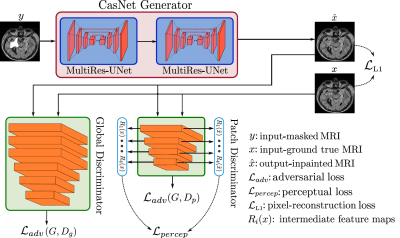
The proposed framework shares the common concept of a deep generative structure as illustrated in Fig. 1. It consists of a generator network (resembling an encoder-decoder structure) and two discriminator networks. The generator is comprised of two MultiRes-UNet6 architectures cascaded together in an end-to-end manner. MultiRes-UNet has an enhanced performance when compared to classical U-nets7 due to the incorporation of high-capacity MultiRes-blocks and intermediate residual paths. The generator receives as input a 2D MRI scan with a randomly cropped arbitrary-shaped mask and is tasked to inpaint the missing region within the masked area. The inpainting performance is guided by the two discriminator networks with different receptive-fields, the global and patch discriminators. These networks are responsible for the calculation of the training loss functions: the adversarial loss, perceptual loss8 and a pixel-reconstruction loss.To examine the performance of the proposed framework, a dataset of T2-weighted (FLAIR) brain MR scans from 44 anonymized volunteers was acquired using a 3T scanner. The data was resampled from an original resolution of 0.5x0.5x1 mm3 to 1x1x1 mm3. Two-dimensional slices were extracted and scaled to 256x256 pixels. A set of 150 masks were utilized to create a paired dataset consisting of incomplete masked-MR and their corresponding ground-truth counterparts. Each mask consisted of a single connected region of arbitrary shape and a random location. For training, 100 masks were utilized on scans from 33 volunteers (≈ 3000 images). The remaining 50 masks were used to test the generalization capabilities of the proposed framework to never-before-seen shapes by creating the validation dataset from the remaining 11 volunteers (≈ 1000 images).
Quantitative and qualitative comparisons were conducted against another generative inpainting approach, the gated convolutions framework9, and the pix2pix image translation network10 to illustrate the performance of the proposed method. All networks were trained for 100 epochs using the ADAM optimizer11 and utilizing their original recommended hyperparameter settings. The resultant images were evaluated using the mean square error (MSE), peak-signal-to-noise ratio (PSNR) and the universal quality index (UQI) as metrics.
Results and Discussion
Quantitative and qualitative results for the proposed IPA-MedGAN framework are illustrated in Table 1 and Fig. 2, respectively. The gated convolutions inpainting framework led to the worst inpainting images. The resultant inpainted regions exhibited a lack of texture clarity with significant changes to the anatomical characteristics when compared to the ground-truth target images. This may be attributed to the fact that this framework was designed for the inpainting of natural images. However, the inpainting of MR images is considerably more challenging due to highly detailed grey matter structures in the brain. The pix2pix image translation network resulted in enhanced quantitative performance. Despite this, when compared to the reference images, the regions inpainted by pix2pix exhibited blurrily details which did not fit homogeneously with the surrounding regions. The proposed IPA-MedGAN framework outperformed the other approaches both qualitatively and quantitatively. From a qualitative perspective, the resultant inpainted regions exhibited detailed textures with more realistic anatomical characteristics. Qunaitiatively, the resultant MSE score has improved to 24.9 compared to a second-best score of 43.77 by pix2pix.Despite the positive results, this study is not without limitations. In the future, we would like to test the inpainting performance on different body regions with more challenging masks representing disjointed regions covering larger areas of the input images. More importantly, we would like to test the impact of inpainting MR images on further post-processing tasks, such as therapy dose calculation and PET-MR attenuation correction. Furthermore, expanding the proposed framework for 3D MR inputs will be investigated.
Conclusion
In this work, we proposed a new generative deep learning framework for the inpainting of missing or distorted regions in brain MR scans. The highly obtained evaluation metrics and detailed qualitative results have indicated the enhanced performance of the framework when compared to prior inpainting networks and translation approaches. The potential of inpainted MR images to enhance the performance of further post-processing tasks will be investigated in the future.Acknowledgements
No acknowledgement found.References
1: K.M. Koch, B.A. Hargreaves, K. Butts Pauly, W. Chen, G.E.Gold, and K.F. King, “Magnetic resonance imaging near metal implants,” Journal of Magnetic Resonance Imaging, vol. 32, no. 4, pp. 773–787, 2010.
2: M. Bertalmio, L. Vese, G. Sapiro, and S. Osher, “Simultaneous structure and texture image inpainting,” IEEE Transactions on Image Processing, vol. 12, no. 8, pp. 882–889, 2003.
3: I. J. Goodfellow, J. Pouget-Abadie, M. Mirza, B. Xu, D. Warde-Farley, S. Ozair, A. Courville, and Y. Bengio, "Generative Adversarial Networks", in Conference on Neural Information Processing Systems, 2014.
4: K. Armanious, S. Gatidis, K. Nikolaou, B. Yang, andT. Kustner, “Retrospective correction of rigid and non-rigid MR motion artifacts using gans,” in IEEE 16th InternationalSymposium on Biomedical Imaging (ISBI), April 2019, pp. 1550–1554.
5: K. Armanious, C. Yang, M. Fischer, T. K¨ustner,K. Nikolaou, S. Gatidis, and B. Yang, “MedGAN: Medical image translation using GANs,” arXiv preprint, 2018.
6: N. Ibtehaz and M. S. Rahman, “MultiResUNet : Rethinking the U-Net architecture for multimodal biomedical image segmentation,” in Neural Networks, vol. 121, pp. 74 – 87, 2020.
7: O. Ronneberger, P. Fischer, and T. Brox, “U-net: Convolutional networks for biomedical image segmentation,” in Medical Image Computing and Computer-AssistedIntervention (MICCAI), vol. 9351, pp. 234–241, 2015.
8: C. Wang, C. Xu, C. Wang, and D. Tao, “Perceptual adversarial networks for image-to-image transformation,” in IEEE Transactions on Image Processing, vol. 27, 2018.
9: J. Yu, Z. Lin, J. Yang, X. Shen, X. Lu, and T. S Huang, “Free-form image inpainting with gated convolution,” arXiv preprint, 2018.
10: P. Isola, J. Zhu, T. Zhou, and A. A. Efros, “Image-to-image translation with conditional adversarial networks,” in Conference on Computer Vision and Pattern Recognition, pp. 5967–5976, 2016.
11: D. P. Kingma, J. Ba, "Adam: A method for stochastic optimization.", in International Conference for Learning Representations, 2015.
Figures