3544
Data Augmentation with Conditional Generative Adversarial Networks for Improved Medical Image Segmentation1Medical Biophysics, University of Toronto, Toronto, ON, Canada, 2Physical Sciences Platform, Sunnybrook Research Institute, Toronto, ON, Canada, 3Institute of Biomaterials and Biomedical Engineering, University of Toronto, Toronto, ON, Canada, 4Computer Science and Medicine, University of Toronto, Toronto, ON, Canada
Synopsis
Performance of machine learning models for medical image segmentation is often hindered by a lack of labeled training data. We present a method for data augmentation wherein additional training examples are synthesized using a conditional generative adversarial network (cGAN) conditioned on a ground truth segmentation mask. The mask is later used as a label during the segmentation task. Using a dataset of N=48 T2-weighted MR volumes of the prostate, our results demonstrate the mean DSC score of a U-Net prostate segmentation model increased from 0.74 to 0.76 when synthetic training images are included with real data.
Introduction
Segmentation of medical images is a crucial step in many clinical and diagnostic procedures1,2. Manual segmentation is the gold-standard, however, automated segmentation via machine learning is desirable to alleviate clinician time and avoid inter-observer variability3. Convolutional neural networks have been shown to be well-suited for segmentation tasks, but require large amounts of labeled training data. Numerous factors affect the availability of labeled data, including patient consent and image acquisition differences between institutions. Data augmentation techniques, which improve the diversity of a dataset, are widely used but many standard techniques are unsuitable for medical imaging applications and generally do not reflect inter-patient variability. We present a novel method for data augmentation using a conditional generative adversarial network (cGAN), wherein the cGAN is conditioned on a segmentation mask derived from an atlas to generate additional, realistic, labeled training examples. Robust data augmentation techniques for the creation of labeled synthetic data would aid segmentation tasks where the set of available training examples is too small to acquire accurate results. In this work, we show that the real data combined with the cGAN generated synthetic data outperforms the non-augmented dataset on the task of prostate segmentation using a U-Net4.Method
Data: Transverse T2-weighted volumes of the prostate (resolution (0.625x0.625x3.6)mm, average of 20 slices per volume) of N=48 prostate patients and accompanying ground truth prostate segmentations from the Medical Segmentation Decathlon 2018 dataset5 were used. The volumes were resampled to an isotropic voxel size of (1.0x1.0x1.0)mm3, to obtain a matrix size of 256x256 for network input, increasing the average number of slices per volume to 70. The 48 volumes were split into four distinct groups, N=32 for generating synthetic images (N=25 for training, N=7 for testing), and N=16 for the segmentation task (N=13 for training, N=3 for testing).Models: A U-Net4 was used for the segmentation task. The U-Net is a fully convolutional autoencoder network featuring skip connections to provide context to the upsampling layers, specifically developed for biomedical image segmentation. The “Pix2Pix” cGAN architecture6 was used for the synthetic image generation task. The model consists of two competing networks–(1) a generator (U-Net) which generates candidate images based on a condition and model distribution and (2) a discriminator (fully convolutional network) that discriminates between the candidate and ground truth images.
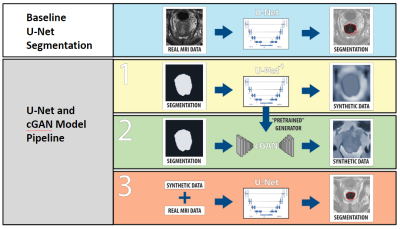
Procedure: The workflow is described in figure 1. The U-Net which is used in the cGAN architecture for image synthesis is independently pre-trained to generate prostate MRIs based on a mask of the prostate. The discriminator component of the cGAN was then added to “fine-tune” the synthetic images to add realism so that synthetic images closer matched the distribution of real images. The synthetic images were then combined with the dataset of real images set aside for the segmentation task and used to train a distinct U-Net for prostate segmentation. A U-Net trained without augmentation techniques (only the real data set) was used as a baseline comparison.
Results
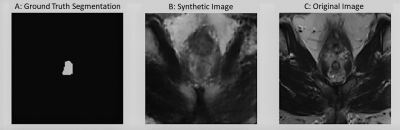
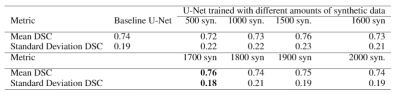
The Dice Similarity Coefficient (DSC) was used for evaluation7. For the baseline U-Net using only the real data, the mean DSC for each volume was 0.74 +/- 0.19. In Table 1, the results are provided for the baseline compared to a U-Net trained on different amounts of synthetic images. With 1700 synthetic training examples, an optimal mean DSC of 0.76 +/- 0.18 standard deviation was achieved (p-value<0.05 found using a Mann-Whitney U test). Figure 4 shows representative samples comparing the baseline U-Net to the augmented U-Net. Figure 5. demonstrates that augmentation using synthetic data increases the number of slices with DSC of 0.8 or greater.Discussion
We have demonstrated that using a cGAN-based data augmentation technique was effective at improving the DSC score of a U-Net trained for prostate segmentation. No additional augmentation techniques were used in order to isolate the effect of our proposed technique. We tentatively attribute the decrease of segmentation performance when 500 synthetic slices were included in the training due to a lack of realism in the synthetic images, which remain differentiable from real data by a human observer. We hypothesize that optimizing the amount of generator pre-training will increase the realism of the synthetic images and improve segmentation performance. The authors acknowledge training the augmented model on synthetic data generated from volumes not included in the training set of the baseline model could lead to skewed results. This issue will be addressed in future work. Future work also consists of determining how increases in performance due to our method compare to other augmentation techniques, the effect of implementing our proposed technique in tandem with other techniques, and duplication of our presented results using other baseline segmentation models and other segmentation tasks to assess the generalizability of the procedure.Conclusion
We presented a data augmentation technique to increase the performance of deep learning-based medical image segmentation models. The technique involves the generation of realistic labeled synthetic data. The average dice score of segmentation model trained on the augmented dataset was improved compared to that of the same model trained on the non-augmented dataset. This technique is expected to help address the need for large amounts of difficult to acquire labeled training data to train deep segmentation models.Acknowledgements
We would like to thank Dr. Anne Martel and the Martel Lab for computational resources. We can also claim no conflicts of interest in this research.References
1. Deklerck, R., Cornelis, J., & Bister, M. (1993). Segmentation of medical images. Image and Vision Computing, 11(8), 486–503. https://doi.org/10.1016/0262-8856(93)90068-R
2. Pham, D. L., Xu, C., & Prince, J. L. (2000). Current Methods in Medical Image Segmentation. Annual Review of Biomedical Engineering, 2(1), 315–337. https://doi.org/10.1146/annurev.bioeng.2.1.315
3. Warfield, S. K., Zou, K. H., & Wells, W. M. (2004). Simultaneous truth and performance level estimation (STAPLE): An algorithm for the validation of image segmentation. IEEE Transactions on Medical Imaging, 23(7), 903–921. https://doi.org/10.1109/TMI.2004.828354
4. Ronneberger, O., Fischer, P., & Brox, T. (2015, October). U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical image computing and computer-assisted intervention (pp. 234-241). Springer, Cham. https://doi.org/10.1007/978-3-319-24574-4_28
5. Simpson, A. L., Antonelli, M., Bakas, S., Bilello, M., Farahani, K., van Ginneken, B., … Cardoso, M. J. (2019). A large annotated medical image dataset for the development and evaluation of segmentation algorithms. Retrieved from http://arxiv.org/abs/1902.09063
6. Isola, P., Zhu, J. Y., Zhou, T., & Efros, A. A. (2017). Image-to-image translation with conditional adversarial networks. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 1125-1134).
7. Zou, K. H., Warfield, S. K., Bharatha, A., Tempany, C. M., Kaus, M. R., Haker, S. J., ... & Kikinis, R. (2004). Statistical validation of image segmentation quality based on a spatial overlap index1: scientific reports. Academic radiology, 11(2), 178-189. https://doi.org/10.1016/S1076-6332(03)00671-8
Figures