3543
Enhanced One-minute EPImix Brain MRI Scans with distortion correction Based on supervised GAN model1Tsinghua University, Beijing, China, 2The Johns Hopkins University, Baltimore, MD, United States, 3Subtle Medical Inc., Menlo Park, CA, United States, 4Karolinska Institutet, Stockholm, Sweden, 5Stanford University, Stanford, CA, United States
Synopsis
EPImix is a one-minute full brain magnetic resonance exam using a multicontrast echo-planar imaging (EPI) sequence, which can generate six contrasts at a time. However, the low resolution and signal-to-noise ratio impeded its application. And EPI distortion makes it harder to improve the quality of imaging. In this study, we applied topup for distortion correction and propose a supervised deep learning model to enhance the EPImix images. We test the GRE, T2, T2FLAIR images on network and analyze the peak signal to noise ratio and structural similarity results. The results suggest that the proposed method can effectively enhance EPImix images.
Introduction
EPImix is a one-minute full brain magnetic resonance exam using a multi-contrast echo-planar imaging (EPI) sequence, which can generate six contrasts at a time1. Due to its high speed acquisition, it can improve patients’ experience and reduce the cost of MRI. However, the low resolution and signal-to-noise ratio (SNR) impeded its wide application. And intrinsic EPI distortion makes it harder to improve the quality of imaging. Fsl-topup is a method that uses to find the susceptibility off-resonance field2. It uses two acquisitions where the acquisition parameters are different so that the mapping between field and distortions are different. In this study, we apply the phase encoding inverse pairs to fsl-Topup and propose a supervised deep learning model. We test the GRE, T2, T2FLAIR images on the network and analyze the peak signal to noise ratio (PSNR) and structural similarity (SSIM) results.Method
1) Dataset89 patients were recruited from Stanford Imaging Hospital, CA, US. All scans were performed at 3T MR system (GE Healthcare). The example of the dataset is shown in Figure 1. The MR protocol included 6 EPIMix sequences (T1‐FLAIR, T2‐w, diffusion‐weighted (DWI), apparent diffusion coefficient (ADC), T*w, T2‐FLAIR) and three sequences (GRE, T2, T2-FLAIR) were scanned in traditional way. The EPImix raw data includes the phase encoding inverse pairs data. EPIMix images are of 256 × 256 pixels, with pixel spacing of 0.9375 mm× 0.9375 mm. Standard images are of 512 × 512 pixels, with pixel spacing of 0.4688 mm× 0.4688 mm. 69 cases are randomly chosen testing and others for testing.
2) Data preprocessing
Due to the limitation of the standard dataset, we only select GRE, T2, T2-FLAIR images from EPImix dataset. Fsl-Topup were applied for distortion correction. Due to the time consuming character of Topup, the phase encoding reversed pairs are resized to 90×90 via bicubic to estimate the field. Then enlarge the mid-result so that they can apply for 256×256. All the images were registered to standard GRE images by spm12. Top and bottom 3 slices were removed.
3) Network
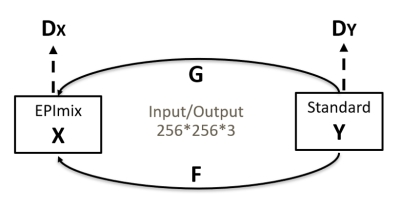
We choose CycleGAN3, shown in Figure 2 as our network. The generators are designed to translate one image style to another so that they can achieve the image enhancement from EPImix to standard images. The discriminators are designed to estimate whether the images are real. The generator is rennet_9block while the discriminator is 70×70 PatchGAN. The inputs of network are in pair. The loss function includes loss LL1, cycle-consistent loss Lcyc, and identity loss Lid . X stands for EPIMix dataset while Y for standard dataset. For the inputs are in pair, we can calculate L1-loss between X and Y. The loss function can be described as: $$L(G,F,D_{X},D_{Y})=L_{Gan}(G,D_{Y},X,Y)+L_{Gan}(F,D_{X},Y,X)+\lambda _{1}L_{cyc}(F,G)+\lambda _{2}L_{id}(F,G)+\lambda _{3}L_{L1}(G)+\lambda _{4}L_{L1}(F)$$
We set λ1 as 10, λ2 as 5, λ3 as 200, λ4 as 200. We trained the network with batch size=10 and instance normalization.
Results and Discussion
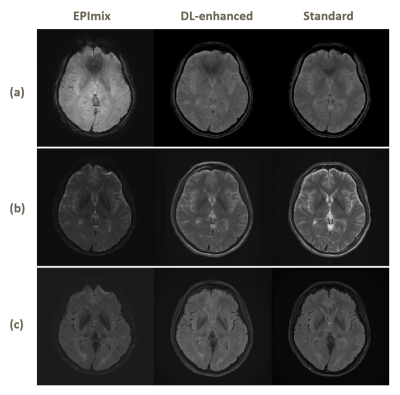
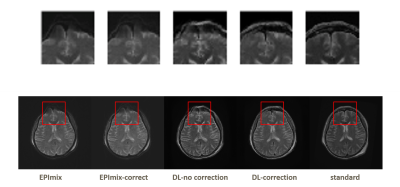
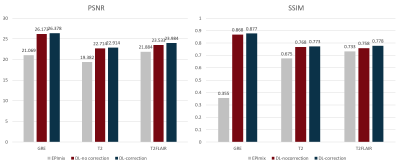
The results are shown in Figure 3. We respectively apply EPImix with/without Topup correction to our neural network to test the effect of Topup. Three channels input share the same patterns and features, so that the details of images can be enhanced, especially in GRE images. The calculated results show that the noise is suppressed and the generated images are more similar to standard images than EPImix images. The correction of distortion only works well on some certain slices that are distorted seriously. As shown in Figure 4, the top of brain and skull have been visibly corrected. The PSNR and SSIM are also significantly improved as shown in Figure 5.The traditional CycleGan model for image style transfer is always unsupervised. Due to well registration and distortion correction, EPImix images are matching with standard images and enable us to do further quality analysis. Moreover, three-channel input/output share the information and generate three contrasts at a time, which are more efficient and accurate. Even though PSNR and SSIM are higher than EPImix images, reader study is desired to test the pathological performance of results. In order to get better registration, more study will focus on 3D sequence of high quality images.
Conclusion
In this study, we use a supervised deep learning network to enhance the EPImix images. The PSNR and SSIM results evidence that the proposed method can effectively improve the visual quality of one-minute EPImix images. This is the benefit to patients’ experience improvement and clinical application of EPImix.Acknowledgements
References
[1] Skare, S., Sprenger, T., Norbeck, O., Rydén, H., Blomberg, L., Avventi, E., & Engström, M. (2018). A 1‐minute full brain MR exam using a multicontrast EPI sequence. Magnetic Resonance in Medicine, 79(6), 3045-3054.
[2] J.L.R. Andersson, S. Skare, J. Ashburner. (2003). How to correct susceptibility distortions in spin-echo echo-planar images: application to diffusion tensor imaging. NeuroImage, 20(2):870-888.
[3] Zhu, J. Y., Park, T., Isola, P., & Efros, A. A. (2017). Unpaired image-to-image translation using cycle-consistent adversarial networks. arXiv preprint.
Figures