3540
Super-Resolution with Conditional-GAN for MR Brain Images
Alessandro Sciarra1,2, Max Dünnwald1,3, Hendrik Mattern2, Oliver Speck2,4,5,6, and Steffen Oeltze-Jafra1,4
1MedDigit, Department of Neurology, Medical Faculty, Otto von Guericke University, Magdeburg, Germany, 2BMMR, Biomedical Magnetic Resonance, Otto von Guericke University, Magdeburg, Germany, 3Faculty of Computer Science, Otto von Guericke University, Magdeburg, Germany, 4Center for Behavioral Brain Sciences (CBBS), Magdeburg, Germany, 5German Center for Neurodegenerative Disease, Magdeburg, Germany, 6Leibniz Institute for Neurobiology, Magdeburg, Germany
1MedDigit, Department of Neurology, Medical Faculty, Otto von Guericke University, Magdeburg, Germany, 2BMMR, Biomedical Magnetic Resonance, Otto von Guericke University, Magdeburg, Germany, 3Faculty of Computer Science, Otto von Guericke University, Magdeburg, Germany, 4Center for Behavioral Brain Sciences (CBBS), Magdeburg, Germany, 5German Center for Neurodegenerative Disease, Magdeburg, Germany, 6Leibniz Institute for Neurobiology, Magdeburg, Germany
Synopsis
In clinical routine acquisitions, the resolution in the slice direction is often worse than the in-plane resolution. Super-resolution techniques can help to retrieve the lack of information. Employing a conditional generative adversarial network (c-GAN), known as pix2pix for T1-w brain images, we were able to reconstruct downsampled images till 10-fold. The neural network was compared with the traditional bivariate interpolation method, and the results show that pix2pix is a valid alternative.
Introduction
Magnetic Resonance Imaging (MRI) is a powerful medical imaging technique. However, several physical, technological and economical aspects limit the spatial resolution achieved in clinical routine acquisitions. Hence, it is common to perform interpolation using bicubic, B-spline and other methods to achieve higher levels of detail1-4. Classical interpolation methods can strongly affect the accuracy of subsequent processing steps such as segmentation or registration5. In this work, we investigate an alternative way to obtain images with higher resolution, making use of a deep learning method, in particular, a conditional Generative Adversarial Network (c-GAN), known as pix2pix6 and compare it with the traditional bivariate spline interpolation method.Methods
The dataset utilized in this work is composed of images acquired with a 7T whole body MRI scanner (Siemens Healthineers, Erlangen, Germany), using a 32-channel head coil (Nova Medical, Wilmington, MA, USA) and an optical motion tracking system (OMTS). The OMTS consisted of a MR compatible camera (MT384i, Metria Innovation Inc., Milwaukee, WI, USA) and a Moiré Phase Tracking (MPT) marker. T1-weighted images were acquired using a 3D-MPRAGE sequence with an isotropic resolution of 0.45 mm3. 21 healthy subjects were scanned. The data were then, downsampled using FreeSurfer7(tri-linear interpolation) in order to obtain 2D paired image samples characterized as follows: original in-plane resolution image paired with the same image downsampled to 1.00x0.45, 2.00x0.45 up to 5.00x0.45 mm2 (Fig. 1). Next, the dataset was split into 17500/1000/400 paired samples for training, validation and testing, respectively. The architecture of pix2pix was set to a Res-Net8 with 9 blocks as generator and a Patch-GAN6 as discriminator. The Pytorch implementation by Jun-Yan Zhu9 was used as a basis. As traditional interpolation method, the bivariate spline technique implemented in Python10 was employed. The obtained results were comparatively assessed by calculating the mean-squared-error (MSE), first between the downsampled image and the original image, and then, between the ground-truth and the predicted image. Furthermore, after skull-stripping and three tissues segmentation (gray matter, GM, white matter, WM and cerebrospinal fluid, CSF) performed using FSL Software11, a multi-class DICE coefficient12, and volume ratios were calculated. We defined the volume ratio as the ratio between the numbers of pixels of each tissue class, e.g., the number of pixels for GM in the predicted image divided by the number of pixels of GM in the ground-truth image: #pixelGM-pred/#pixelGM-ground-truth. The processing pipeline is shown in Figure 4.Results
Figure 2 and 3 show the results of one sample for each chosen downsampling factor. In Figure 2, the results from the interpolation test using the bivariate spline algorithm and in Figure 3, the results after the application of pix2pix are illustrated. Figure 5 shows three box plots: MSE values, DICE coefficients and the volume ratios (from left to right). The MSE values, as explained above, are calculated first, between input (first, under-sampled and then, upscaled image using Nearest-Neighbour Interpolation) and ground-truth (original resolution) and second, between the interpolated (using B-Splines)/predicted and the original image. The values of the DICE coefficients are the averages across the three classes GM, WM and CSF. The rightmost box plot shows the volume ratios, also, averaged over the three classes.Discussion
Pix2pix yields images less blurred (Fig. 3) than bivariate spline interpolation (Fig. 2), but in the case of a high downsampling factor, not all brain structures match with the original ones (rightmost columns in Fig. 3). Although the bivariate spline interpolation is not able to recover most of the edge information when the downsampling factor is high (rightmost columns in Fig. 2), it does not introduce spurious structures. The quantitative results in Figure 5 suggest favoring the deep learning approach. The MSE values of the predicted images are substantially smaller than the interpolated ones. The blur level is likely to have a substantial impact. The DICE coefficients provide an estimation of the tissue segmentation equality. The values for the predicted images are consistently higher as compared to the values of the corresponding interpolated ones. For the volumetric analysis, images predicted using pix2pix yield ratios closer to 1.0 compared with the ones from the interpolated images. This indicates that the predicted images just like the original images can be used for a volumetric tissue evaluation.Conclusion
Visual comparison and quantitative assessments showed that pix2pix is able to reconstruct images with details that very well match the original ones even for a 10-fold interpolation. While the results are encouraging, their accuracy may be further increased by fine-tuning the network. Especially on details, as well as suppress the wrong predicted brain structures or their overconfident prediction respectively.Acknowledgements
This work was supported by the federal state of Saxony-Anhalt under grant number ‘I 88’ (MedDigit) and by the NIH, grant number 1R01-DA021146.References
- José V. Manjón, Pierrick Coupé, Antonio Buades, Vladimir Fonov, D. Louis Collins, Montserrat Robles, Non-local MRI upsampling, Medical Image Analysis, Volume 14, Issue 6, 2010, Pages 784-792, ISSN 1361-8415, https://doi.org/10.1016/j.media.2010.05.010
- Kashou, N.H., Smith, M.A. & Roberts, C.J. Ameliorating slice gaps in multislice magnetic resonance images: an interpolation scheme. Int J CARS (2015) 10: 19. https://doi.org/10.1007/s11548-014-1002-3
- Bai, Ying & Han, Xiao & Prince, Jerry. (2004). Super-resolution reconstruction of MR brain images. https://pdfs.semanticscholar.org/35ca/497323acad223eb38b79b14bc25c8f352a25.pdf
- José V. Manjón, Pierrick Coupé, Antonio Buades, D. Louis Collins, and Montserrat Robles, “MRI Superresolution Using Self-Similarity and Image Priors,” International Journal of Biomedical Imaging, vol. 2010, Article ID 425891, 11 pages, 2010. https://doi.org/10.1155/2010/425891
- Ivana Despotović, Bart Goossens, and Wilfried Philips, “MRI Segmentation of the Human Brain: Challenges, Methods, and Applications,” Computational and Mathematical Methods in Medicine, vol. 2015, Article ID 450341, 23 pages, 2015. https://doi.org/10.1155/2015/450341.
- Phillip Isola, Jun-Yan Zhu, Tinghui Zhou, Alexei A. Efros. “Image-to-Image Translation with Conditional Adversarial Networks”. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). DOI: 10.1109/CVPR.2017.632, https://arxiv.org/pdf/1611.07004v1.pdf
- FreeSurfer: https://surfer.nmr.mgh.harvard.edu/
- He K. et. al: Deep Residual Learning for Image Recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). DOI: https://doi.org/10.1109/CVPR.2016.90
- Pytorch implementation: https://github.com/junyanz/pytorch-CycleGAN-and-pix2pix/
- Bivariate Spline Interpolation python https://docs.scipy.org/doc/scipy/reference/generated/scipy.interpolate.RectBivariateSpline.html
- FSL Software: https://fsl.fmrib.ox.ac.uk/fsl/fslwiki
- Dice, L. R.: Measures of the Amount of Ecologic Association Between Species. Ecology 1945; 26(3):297-302.
Figures
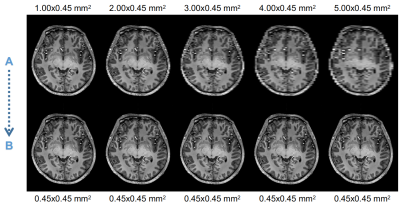
Examples of paired samples used for training pix2pix network. “A”: images downsampled. “B”: reference slice, original resolution.
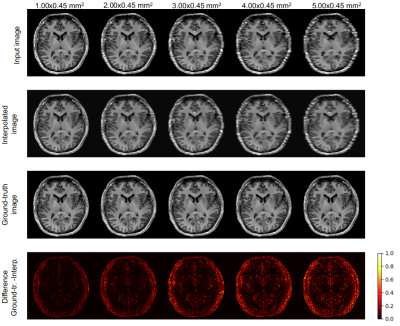
Interpolation of downsampled images.
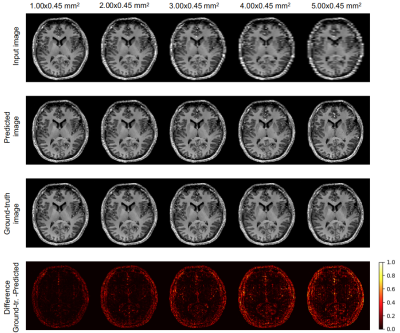
Application of pix2pix on downsampled images.
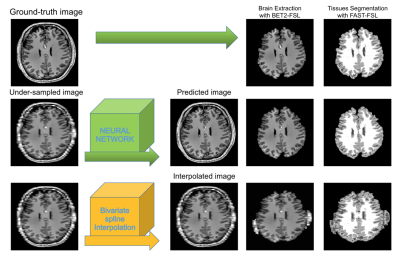
Pipeline used for processing the results.
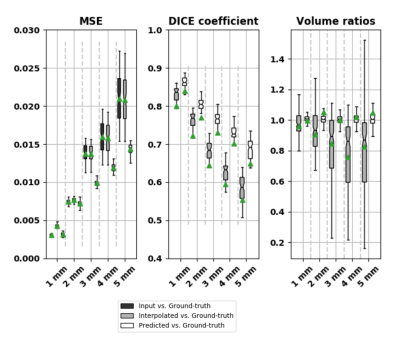
Results: Mean-squared-error (MSE), DICE coefficient and Volume ratios. The green triangles indicate the mean values and the lines the correspondent median values.