3537
USR-Net: A Simple Unsupervised Single-Image Super-Resolution Method for Late Gadolinium Enhancement CMR1Department of Computer Science and Technology, University of Cambridge, Cambridge, United Kingdom, 2Cardiovascular Research Centre, Royal Brompton Hospital, London, United Kingdom, 3National Heart and Lung Institute, Imperial College London, London, United Kingdom
Synopsis
Three-dimensional late gadolinium enhanced (LGE) CMR plays an important role in scar tissue detection in patients with atrial fibrillation. Although high spatial resolution and contiguous coverage lead to a better visualization of the thin-walled left atrium and scar tissues, markedly prolonged scanning time is required for spatial resolution improvement. In this paper, we propose a convolutional neural network based unsupervised super-resolution method, namely USR-Net, to increase the apparent spatial resolution of 3D LGE data without increasing the scanning time. Our USR-Net can achieve robust and comparable performance with state-of-the-art supervised methods which require a large amount of additional training images.
Introduction
Promising results have been reported that late gadolinium enhanced (LGE) CMR is helpful to show native and post-ablation treatment scar within the left atrium (LA) in patients with AF [1]. However, on-going concerns about the accuracy of identifying scars with LGE CMR still exist, partially because the limited spatial resolution of the LGE CMR leads to low diagnostic value due to the very thin LA wall [2,3]. Increasing the acquired spatial resolution of 3D LGE is generally not possible in practice, because it is expensive and time-consuming. Thus, super-resolution (SR) based post-processing becomes a promising option to increase the spatial resolution without increasing acquisition durations of the 3D LGE data. In this study, we propose a slice-wise deep learning based unsupervised SR workflow, namely USR-Net, for LGE CMR.Methods
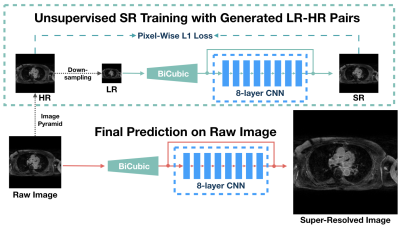
Convolutional neural network (CNN) based single image SR (SISR) methods have been successfully applied on natural images [5-7]. Additionally, generative adversarial networks (GANs) [4], make it possible to achieve higher spatial resolution with high perceptual quality of medical images instead of increasing scanning time [15,17]. However, these supervised methods are limited by: (a) the requirement of a large amount of training data and (b) data variances among patients. Thus, inspired by the idea that inner features of images repeatedly appear in various scales [18], we propose a slice-wise unsupervised SR workflow, USR-Net (Figure 1), to achieve SR on one LGE slice just using its inner features. USR-Net simply consists of a bicubic interpolation layer and a residual CNN with 8 hidden layers. The input low-resolution (LR) image is firstly up-sampled to the target size with bicubic interpolation, then passed to the residual CNN, which aims to predict the difference between the interpolated images and high-resolution (HR) ground truth (GT) images. USR-Net requires no additional training images, but generates LR-HR image pairs via a pyramid architecture for training. Thus, the multi-scale inner features are learned, and then used to predict the super-resolved image of the raw input directly. At last, back-projection [19] is applied as the post-processing for the final SR image. We compared USR-Net with bicubic interpolation and state-of-the-art (SOTA) supervised SISR methods (e.g. SRResNet [5], EDSR [6], and RDN [7]) for simulated X2 and X4 magnification tasks. With ethical approval, 1184 slices from 3D LGE datasets of 20 patients with longstanding persistent AF were collected on a Siemens Magnetom Avanto 1.5T scanner and used for evaluation. During acquisition, an inversion prepared segmented gradient echo sequence ((1.4–1.5)×(1.4–1.5)×4mm3 reconstructed into (0.7–0.75)×(0.7–0.75)×2mm3) were used 15 minutes after gadolinium administration (Gadovist—gadobutrol, 0.1mmol/kg body weight) [10] to perform transverse navigator-gated 3D LGE CMR [8], [9]. To better null the signal from normal myocardium, a dynamic inversion time (TI) was incorporated[11]. CLAWS respiratory motion control was used to increase respiratory efficiency when the CMR data was acquired during free-breathing [2]. To reduce the navigator artefact in the LA, a navigator-restore delay of 100 ms was introduced [8]. In the simulated experiments, we considered the acquired data as HR GT, and we considered the down-sampled (by a factor of 2 or 4) images as raw input of USR-Net. USR-Net was applied with each slice independently, while supervised methods were applied with 5-fold cross validation to acquire super-resolved images of all slices. All SR results are compared with GT and evaluated using Peak-Signal-to-Noise-Ratio (PSNR) and Structural-SIMilarity (SSIM) [13].Results
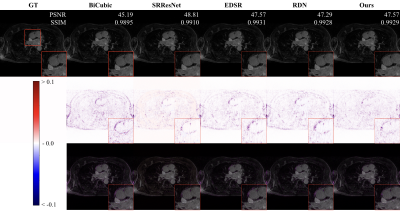
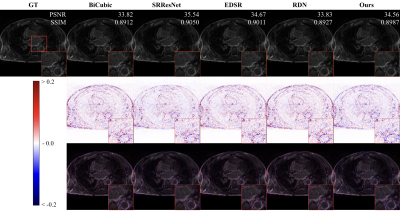
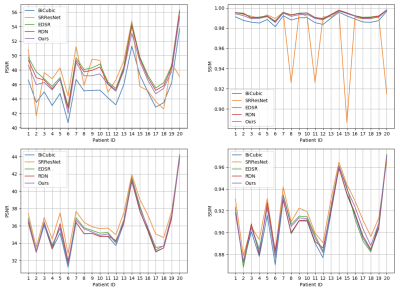
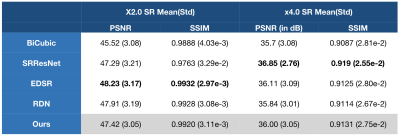
Figure 2 and 3 perceptually show example results of X2 and X4 magnifications. Unlike supervised methods generating more errors around edges, the differences between USR-Net predicted SR images and GT images generally spread over the whole image. Figure 4 illustrates the patient-wise mean PSNR and SSIM of all methods, and Figure 5 shows the mean performance (with standard deviation) of all the 1184 slices. Our USR-Net has achieved robust and comparable performance with supervised methods with both magnification factors.Discussion
CNN based SR methods provide an alternative way to achieve higher spatial resolution of the LGE CMR images without additional scanning time, which leads to improved visualization, quantification and diagnosis of LA scars. With our best experience, this abstract is the first comparison study of SOTA CNN based supervised SISR methods on LEG CMR. From our experiments, we find that supervised methods are limited by the amount of training data, and patient-wise data variances also leads to unstable performance. Our proposed unsupervised SR workflow (USR-Net) solves both issues well. First, all training LR-HR pairs are generated from the raw image, requiring no extra training data. Second, USR-Net aims to learn the inner features of the image itself, meaning no data variance at all. Comparing with bicubic interpolation, USR-Net achieves much better performance on both PSNR and SSIM. More importantly, as an unsupervised method, USR-Net achieves robust and comparable performance with SOTA supervised methods.Conclusion
In this study, our contributions are twofold: first, we reimplement and compare the SOTA SISR methods for 3D LGE CMR images, and illustrate their limitations; second, we propose an unsupervised workflow, i.e., USR-Net, for slice-wised LGE CMR super-resolution, which has achieved comparable performance with supervised methods. Requiring no extra training data, our proposed USR-Net could lead to better visualization and quantification of LGE CMR images with much higher super-resolution.Acknowledgements
Jin Zhu’s PhD research is funded by China Scholarship Council (grant No.201708060173).
Guang Yang is funded by the British Heart Foundation Project Grant (Project Number: PG/16/78/32402).
References
[1] D. C. Peters et al., “Detection of pulmonary vein and left atrial scar after catheter ablation with three-dimensional navigator-gated delayed enhancement MR imaging: initial experience.,” Radiology, vol. 243, no. 3, pp. 690–695, 2007.
[2] C. McGann et al., “Atrial fibrillation ablation outcome is predicted by left atrial remodeling on MRI,” Circ. Arrhythmia Electrophysiol., vol. 7, no. 1, pp. 23–30, 2014.
[3] J. L. Harrison et al., “Repeat Left Atrial Catheter Ablation: Cardiac Magnetic Resonance Prediction of Endocardial Voltage and Gaps in Ablation Lesion Sets,” Circ. Arrhythmia Electrophysiol., vol. 8, no. 2, pp. 270–278, Apr. 2015.
[4] I. Goodfellow, J. Pouget-Abadie, and M. Mirza, “Generative Adversarial Networks,” arXiv Prepr. arXiv …, pp. 1–9, 2014.
[5] C. Ledig et al., “Photo-Realistic Single Image Super-Resolution Using a Generative Adversarial Network,” 2016.
[6] Lim, Bee, et al. "Enhanced deep residual networks for single image super-resolution." Proceedings of the IEEE conference on computer vision and pattern recognition workshops. 2017.
[7] Zhang, Yulun, et al. "Residual dense network for image super-resolution." Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2018.
[8] D. C. Peters et al., “Recurrence of Atrial Fibrillation Correlates With the Extent of Post-Procedural Late Gadolinium Enhancement. A Pilot Study,” JACC Cardiovasc. Imaging, vol. 2, no. 3, pp. 308–316, 2009.
[9] R. S. Oakes et al., “Detection and quantification of left atrial structural remodeling with delayed-enhancement magnetic resonance imaging in patients with atrial fibrillation,” Circulation, vol. 119, no. 13, pp. 1758–1767, 2009.
[10] J. Keegan, P. Jhooti, S. V. Babu-Narayan, P. Drivas, S. Ernst, and D. N. Firmin, “Improved respiratory efficiency of 3D late gadolinium enhancement imaging using the continuously adaptive windowing strategy (CLAWS),” Magn. Reson. Med., vol. 71, no. 3, pp. 1064–1074, 2014.
[11] J. Keegan, P. D. Gatehouse, S. Haldar, R. Wage, S. V Babu-Narayan, and D. N. Firmin, “Dynamic inversion time for improved 3D late gadolinium enhancement imaging in patients with atrial fibrillation.,” Magn. Reson. Med., vol. 73, no. 2, pp. 646–54, Feb. 2015.
[12] J. Keegan, P. Drivas, and D. N. Firmin, “Navigator artifact reduction in three-dimensional late gadolinium enhancement imaging of the atria,” Magn. Reson. Med., vol. 785, pp. 779–785, 2013.
[13] C.-Y. Yang, C. Ma, and M. Yang, “Single-Image Super-Resolution: A Benchmark,” in Computer Vision – ECCV 2014, 2014, pp. 372–386.
[14] M. Seitzer et al., “Adversarial and Perceptual Refinement for Compressed Sensing MRI Reconstruction,” in Medical Image Computing and Computer Assisted Intervention (MICCAI 2018), 2018.
[15] Jin Zhu, Guang Yang, and Pietro Lio. "Lesion focused super-resolution." Medical Imaging 2019: Image Processing. Vol. 10949. International Society for Optics and Photonics, 2019.
[16] G. Yang et al., “Fully Automatic Segmentation and Objective Assessment of Atrial Scars for Longstanding Persistent Atrial Fibrillation Patients Using Late Gadolinium-Enhanced MRI,” Med. Phys., vol. 45, no. 4, pp. 1562–1576, 2018.
[17] Jin Zhu, Guang Yang, and Pietro Lio. "How Can We Make GAN Perform Better in Single Medical Image Super-Resolution? A Lesion Focused Multi-Scale Approach." arXiv preprint arXiv:1901.03419 (2019).
[18] Assaf Shocher, Nadav Cohen, and Michal Irani. "“Zero-Shot” Super-Resolution using Deep Internal Learning." Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2018.
[19] Irani, Michal, and Shmuel Peleg. "Improving resolution by image registration." CVGIP: Graphical models and image processing 53.3 (1991): 231-239.
Figures