3536
Deep PET-Prior: MR-derived Zero-Dose PET Prior for Differential Contrast Enhancement of PET1Department of Radiology and Biomedical Imaging, University of California, San Francisco, San Francisco, CA, United States
Synopsis
We introduce a deep neural network scheme for predicting FDG-PET activity from a T1-weighted MR volume. This is useful for creating realistic anatomy-conforming synthetic PET data for prototyping of PET reconstruction algorithms, e.g. from abundant MR-only exam data. While deep networks can learn the average or nominal uptake patterns, in most cases, MR is ultimately incapable of fully predicting PET activity due to fundamental differences in the sensing modalities. We show, however, that these MR-derived “zero-dose” images can aid in differential contrast enhancement and visualization of PET by localizing and highlighting activity uniquely detected by the PET radiotracers.
Introduction
Joint PET/MR imaging is gaining popularity in clinical practice, with many case studies indicating improved diagnostic capabilities by fusing the two modalities. While MR is currently used in PET image reconstruction as a MR-derived attenuation correction (pseudo-CT) [1], or directly in joint estimation of attenuation and PET activity [2], PET data is typically viewed as an additional layer of information atop MR-derived anatomical- or contrast-imagery. However, in many cases the resolution of PET is substantially lower than that of MRI, with diagnostic images rendered with voxels 2-3 times larger than that of typical MR imagery. At this resolution, radiologists must be careful to separate image artifacts and blurring of anatomy from pathology.In this vein, generation of large, realistic datasets of anatomy-conforming synthetic PET imagery is important for improving the next generation of PET/MR reconstruction algorithms in the presence of pathology, irregular anatomy, and implants [3]. Unlike labor-intensive atlas-based techniques, deep neural networks offer a way to implicitly complete this mapping directly from a database of patients’ MRI and PET volumes [4]. Still MR-derived techniques are fundamentally limited in their ability to predict PET-tracer concentration from MRI intensity or texture alone, instead only learning nominal or “average” uptake patterns.
Despite a mismatch in the modalities, we positively exploit this limitation by deliberately trying to predict PET activity from MR activity using a deep neural network. When supplied with an MR volume, the network produces a “zero-dose” PET volume prediction that is used to enhance the contrast of the real PET volume. Qualitatively, contrast enhanced PET images visually indicate the location of increased or decreased activity relative to the MRI-derived baseline. Quantitatively, the mutual information between the contrast enhanced PET and the MRI is reduced, compared to the mutual information of the original PET and MRI. Thus, the proposed zero-dose contrast-enhanced PET imagery provides information that is quantitatively different from MRI. We evaluate this methodology on a dataset of whole-body PET/MRI images (PET images and T1-weighted MRI) and qualitatively describe the features of such contrast enhanced imagery. We suggest potential avenues for utilizing these to characterize pathology.
Methods
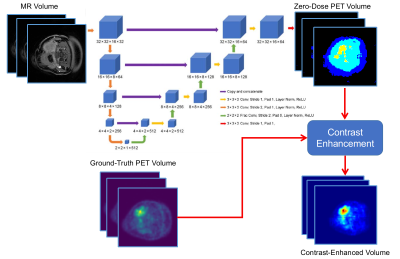
Dataset: A dataset of 58 clinical whole-body PET-MRI exams, collected using a GE Signa PET/MRI system, were selected for retrospective study. The MRI imagery are LAVA-FLEX water images from a two-point Dixon spoiled gradient echo sequence. The PET radiotracer was fluorodeoxyglucose (FDG) for all the studies.Neural Network for Domain Translation: A 3D convolutional deep neural network (U-Net) network [5] was used for predicting co-registered “zero-dose” PET activity volumes from T1-weighted MRI volumes. The network architecture is shown in Figure 1. The network was trained using standard mean absolute error loss using the Adam optimizer with a learning rate of 0.001.
Differential Contrast Enhancement: We create differential contrast-enhanced PET imagery by comparing the real PET volume with the aforementioned MRI-derived zero-dose PET activity prediction. In addition to absolute and relative differences, normalized zero-dose activity images were used, as:
$$\text{PET}_{\text{CE-relative}} = \Big(\frac{ |\hat{y}|}{|\epsilon + \hat{y}|}\Big) \odot y$$
where $$$y$$$ is the true PET volume, $$$\hat{y}$$$ is the MR-derived zero-dose PET volume, and $$$\odot$$$ is the Hadamard product.
Mutual information as a measure of redundancy: We use mutual information to measure the amount of information shared (statistical correlation) between PET/MR images, and show that zero-dose DCE PET imagery provides quantitatively different information than MR images. For simplicity, we compute these over axial slices of the co-registered PET/MR volumes using intensity histograms, and report changes in the average normalized ratio.
Results
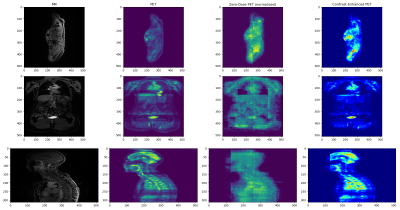
Figure 2 shows a comparison of the MR, PET, zero-dose PET, and contrast-enhanced zero-dose PET for one test subject. Despite the obvious blocking artifacts in the predicted zero-dose PET image, the contrast-enhanced imagery exhibits excellent localization of PET activity, while minimizing display of nominal uptake patterns throughout the body. Quantitatively, the mutual information between the contrast-enhanced volumes and the MR volumes were substantially lower than the mutual information between the MR volume and the original PET volume, indicating that the contrast-enhanced imagery contains information fundamentally different than that of the MR imagery. Although explicit penalization using the approximate KL divergence of the PET volume and zero-dose PET volume was considered, the training scheme was unstable; this should be considered in future work. Still, using the relative zero-dose contrast enhancement resulted in an average of 24% reduction in the mutual information between PET and MR imagery, as measured on a testing dataset of 28 whole-body co-registered PET/MR exams.Conclusions
We proposed a method for predicting zero-dose PET volumes from MR volumes using deep learning. The resulting zero-dose imagery may be used to improve the contrast and highlight unique tracer-specific localization information from real (full- or low-dose) PET imagery by removing nominal uptake patterns. These images may serve as a clinical tool for characterization of abnormal uptake patterns or pathology.Acknowledgements
This work was supported by NIH NCI grant number R01CA212148.References
[1] Leynes, A. P., Yang, J., Wiesinger, F., Kaushik, S. S., Shanbhag, D. D., Seo, Y., ... & Larson, P. E. (2017). Direct pseudoCT generation for pelvis PET/MRI attenuation correction using deep convolutional neural networks with multi-parametric MRI: zero echo-time and dixon deep pseudoCT (ZeDD-CT). Journal of Nuclear Medicine, jnumed-117.
[2] Bal, G., Kehren, F., Michel, C., Watson, C., Manthey, D., & Nuyts, J. (2011). Clinical evaluation of MLAA for MR-PET. Journal of Nuclear Medicine, 52(supplement 1), 263-263.
[3] Arbib, M. A., Bischoff, A., Fagg, A. H., & Grafton, S. T. (1994). Synthetic PET: Analyzing large‐scale properties of neural networks. Human Brain Mapping, 2(4), 225-233.
[4] Xiang, L., Qiao, Y., Nie, D., An, L., Lin, W., Wang, Q., & Shen, D. (2017). Deep auto-context convolutional neural networks for standard-dose PET image estimation from low-dose PET/MRI. Neurocomputing, 267, 406-416.
[5] Ronneberger, O., Fischer, P., & Brox, T. (2015, October). U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical image computing and computer-assisted intervention (pp. 234-241). Springer, Cham.
Figures