3534
Weakly Supervised Exclusion of Non-Tumoral Enhancement in Low Volume Dataset for Breast Tumor Segmentation1Radiology, Columbia University, New York, NY, United States, 2Stony Brook University, Stony Brook, NY, United States
Synopsis
Quantitative measures of breast functional tumor volume are important response predictors of breast cancer undergoing chemotherapy. Automated segmentation networks have difficulty excluding non tumoral enhancing structures from their segmentations. Using a small small DCE-MRI dataset with coarse slice level labels to weakly supervised segmentation was able to exclude large portions of non tumor structures. Without manual pixel wise segmentation, our Class activation map based region proposer excluded 67% of non-tumoral voxels in a sagittal slice from downstream segmentation networks while maintaining 94% sensitivity.
Introduction
Background parenchymal enhancement (BPE) on Dynamic Contrast Enhanced MRI (DCE-MRI) may obscure or mimic malignancy 1 .Measures of breast tumor volume are important response predictors of pathologic response to chemotherapy2. Breast tumor volumes are often measured by thresholding enhancement ratio in DCE-MRI(3). This methodology is not reliable in patients that exhibit strong BPE as the parenchymal enhancing tissue may exceed threshold enhancement ratio and be included in tumor volume measurements.Deep learning models have become very popular due to their simplicity of use, but require large quantities of data to train. High quality labeled datasets are sparse in medical imaging due to the tedious task of manual supervised annotations by human experts. Methods to reduce this expert supervision required for deep learning based segmentation has popularized weakly supervised networks, which utilize large amounts of lower quality labeled data. These networks can use small datasets to initialize downstream networks with domain expertise or constrain downstream networks within reasonable bounds. We propose a weakly supervised network to initialize and constrain a downstream tumor segmentation network from activating on BPE.
Traditional Deep U-Net style networks segment isolated tumors well in DCE-MRI, but perform poorly in areas containing other enhancing structures. We propose training a network with coarse slice level labels to learn tumor related enhancement compared with non tumor BPE. Thresholded Class Activation maps4 will then be used to propose tumor regions in the DCE-MRI dataset. These tumor region proposals will keep the downstream networks from activating on enhancing structures such as the heart, lymph nodes, and background parenchymal enhancement.
Methods
PatientsA HIPPA compliant IRB approved retrospective review of 141 patients with Breast cancer who underwent MRI prior to the initiation of neoadjuvant chemotherapy were identified.
MRI
MRI was performed on 1.5T and 3T GE Signa Excite scanners. Bilateral T1 weighted Fat Saturated 3D FGRE (TR:3.6-13ms, TE:1.1-6.5ms, FA:10-35, BW 26-32 khz) was performed before and after bolus injection of gadobenate dimeglumine (0.1mmol/kg).
Pre-Processing
Pre and two post contrast dynamics were stacked in a 4D Volume (X,Y,Z,T) and intensity normalized to the maximum signal intensity of the stack. The largest dimension in the sagittal plane (AP/SI) was rescaled bilinearly to a matrix size of 256, and the other dimension zero filled to 256. Sagittal slices containing tumor were labeled by non expert using ITK SNAP. Slices were verified to contain tumor by a breast fellowship trained radiologist.
Weak Supervision
A standard Resnet-34 was used for classification of each sagittal slice as tumor or normal. A thresholded class activation map (CAM) is used as a region proposal network and is produced by diverting a forward pass of test images at the last fully convolutional layer and resizing the map to input image dimensions with bilinear interpolation. The CAM is thresholded at 50% and used as a region proposal network within a sagittal slice. Training was performed over 100 epochs at learning rate of 0.00005 with an Adam optimizer and binary cross entropy loss function.
Testing
The testing dataset consisted of 20% of all patients held out from training. Sensitivity, specificity and accuracy performance statistics of the sagittal slice classifier network are analyzed on the holdout testing data. The thresholded CAM is applied as a mask to the test dataset. The region proposals are applied only to slices that contain tumor.Sensitivity, specificity and accuracy performance statistics of the sagittal slice classifier network are analyzed on the holdout testing data.
Results
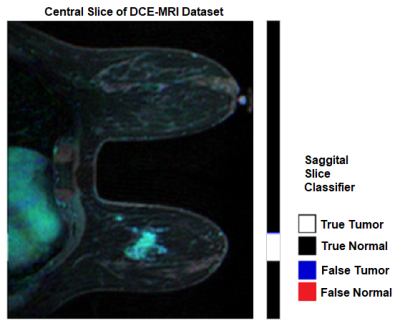
The datasets of 28 patients (20%) are held out from training for use as a testing dataset.Sagittal Slice Classifier
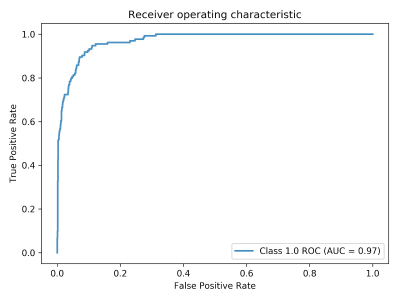
The Sagittal Slice classifier is tested on the 8190 slices of the 28 patient testing dataset. An example of classification result is depicted in figure 1. On the test dataset, the sagittal slice classifier achieved an accuracy of 95.07%, sensitivity of 72.39%, and specificity of 97.40% with an AUC of 0.97 as depicted in figure 2.
CAM Region Proposer
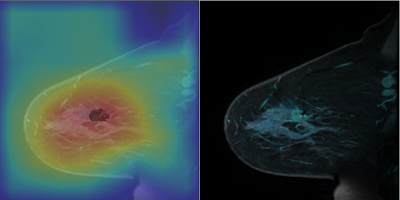
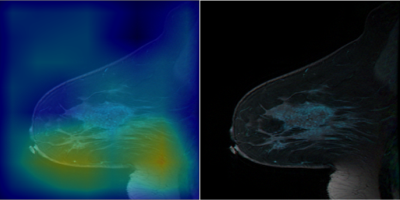
The CAM based region proposer is tested on the 1000 slices of the 28 patient testing dataset that contains tumor. An example of the CAM prior to threshold masking are depicted in figures 3 and 4. Figure 3 shows that the CAM activates in areas of tumor and not BPE and lymph nodes. The CAM also excludes other enhancing structures such as the heart and enhancing vessels. The CAM based region proposer achieves a sensitivity of 94%, specificity 67%, and accuracy 67% over the 1000 test slices.
Discussion/Conclusion
Using non-expert and coarse labeling of data, a CAM based region proposal network excludes a majority of non tumor enhancing structures from downstream segmentation. This network excludes 67% of non tumor voxels while retaining high sensitivity without manual pixel level annotation by an expert. The sagittal slice classifier can learn from the full 4D DCE-MRI dataset and train its vast parameter space without fine data curation. The features learned in this classifier can be transferred to the encoder in a U-Net for segmentation tasks. The parameters sharing in this classifier backbone efficiently populate the deep learning model parameter space similar to Faster-RCNN5 and YOLO6 methods.Acknowledgements
No acknowledgement found.References
1. S.-Y. Kim et al., Effect of Background Parenchymal Enhancement on Pre-Operative Breast Magnetic Resonance Imaging: How It Affects Interpretation and the Role of Second-Look Ultrasound in Patient Management. Ultrasound Med. Biol. 42, 2766–2774 (2016).
2. N. M. Hylton et al., Neoadjuvant Chemotherapy for Breast Cancer: Functional Tumor Volume by MR Imaging Predicts Recurrence-free Survival-Results from the ACRIN 6657/CALGB 150007 I-SPY 1 TRIAL. Radiology. 279, 44–55 (2016).
3. N. M. Hylton, Magn. Reson. Imaging Clin. N. Am., in press.
4. B. Zhou, A. Khosla, A. Lapedriza, A. Oliva, A. Torralba, Learning Deep Features for Discriminative Localization. arXiv [cs.CV] (2015), (available at http://arxiv.org/abs/1512.04150).
5. S. Ren, K. He, R. Girshick, J. Sun, Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. arXiv [cs.CV] (2015), (available at http://arxiv.org/abs/1506.01497).
6. J. Redmon, S. Divvala, R. Girshick, A. Farhadi, You Only Look Once: Unified, Real-Time Object Detection. arXiv [cs.CV] (2015), (available at http://arxiv.org/abs/1506.02640).
Figures