3527
A two-stage deep learning method for the identification of rectal cancer lesions in MR images1China Medical University, Shenyang, China, 2Paul C. Lauterbur Research Center for Biomedical Imaging, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China, 3Cancer Hospital of China Medical University, Shenyang, China, 4Department of Electronic Information Engineering, Nanchang University, Nanchang, China
Synopsis
End-to-end deep learning methods, such as the well-known U-Net, have achieved great successes in biomedical image segmentation tasks. These models are often fed with the full field of view images which may contain irrelevant organs or tissues influencing the segmentation performance. In this study, targeting at the accurate segmentation of rectal cancer lesions in T1-weighted MR images, we propose a two-stage deep learning method that is composed of a detection stage and a segmentation stage. Experimental results show that under the guidance of the detected bounding boxes, better segmentation performance is achieved.
INTRODUCTION
Rectal cancer is one of the most common tumors in the digestive system. As one of the most widely adopted screening tools for rectal cancer diagnosis, magnetic resonance imaging (MRI) plays an important role in the clinic 1. However, manual diagnosis and segmentation of the rectal cancer lesions in the preoperative MRI images, which is very important for the patients who will undergo a surgery or further chemotherapy and radiotherapy, is very tedious and time-consuming for the radiologists 2. It would be of great help to develop a computer-aided system to automatically and accurately locate the lesions. There have been many efforts devoted to develop end-to-end deep learning models for medical image segmentation, such as the famous U-Net 3 and SegNet 4. These methods were designed to directly handle the full field of view (FOV) images or the extracted image patches, which is not appropriate for our task of rectal cancer segmentation. The MR images we have contain a lot of chaos information, namely other organs except for the rectal areas. To this end, we develop a two-stage deep learning method to identify the rectal cancer regions from the full FOV images. Experimental results show that the proposed method can automatically segment the rectal cancer lesions with improved accuracy, which is expected to be further optimized help radiologists diagnose the images and design the following treatment plans.METHODS
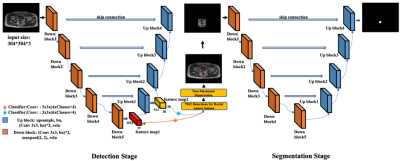
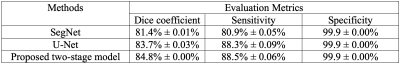
The workflow of our proposed method is shown in Figure 1. Overall, the full FOV images first go through the detection model and bounding boxes containing the lesions are generated. Then, a segmentation model is employed to segment the lesions from the bounded images. Both the detection and the segmentation models are based on the U-Net. Specifically, all of the down sampling blocks in the U-Net and feature maps generated by U-Net down sampling block5 and up sampling block1 constitute the detection branch of our model. The detection strategy is inspired from the single shot multibox detector 5. For the following precise lesion segmentation task, we simply utilize the U-Net in this study as our objective is to investigate the overall framework rather than the specific model architectures. The final segmentation performance of our proposed method is compared to those of the two widely applied end-to-end deep learning segmentation models, U-Net and SegNet. For all the experiments, we use a dice loss and a learning rate of 10-4.We evaluate our proposed model and the comparison networks on a dataset composed of 1392 T1-weighted MRI image slices from 192 patients who have been diagnosed with rectal cancer. We conduct 2D image segmentation instead of 3D as the slice depth is too large compared to the 2D image resolutions. To avoid possible bias, it is guaranteed that slices from the same patient go into either the training dataset or the testing dataset. Five-fold cross-validation experiments were conducted. All the experiments were repeated for three times with different initializations to ensure the reproducibility of the observations. Dice similarity coefficient, sensitivity, and specificity were calculated to characterize the segmentation performances of the different models 6-8.RESULTS
Detailed quantitative results are listed in Table 1. It can be observed that for the two baseline models, U-Net is consistently better than SegNet. With the help from the first-stage detection model, the segmentation performance of U-Net is further improved.Figure 2 shows several examples of the segmentation results. The three examples clearly show that our proposed first-stage detection model can accurately find the bounding boxes containing the lesion regions. Moreover, based on the detected bounding boxes, our second-stage segmentation model can more precisely define the lesion boundaries.
DISCUSSION
Our results on the clinic dataset confirm the feasibility of the proposed two-stage workflow. The first segmentation example in Figure 2 indicates that the detection stage can help get rid of the interference from the irrelevant structures and reduce the false-positive segmentation results. On the other hand, the third example in Figure 2 reflects that with the bounded inputs, our segmentation model can focus more on the important regions and locate the lesions with higher precision.CONCLUSION
We proposed a two-stage deep learning method to achieve accurate rectal cancer lesion segmentation. The method was realized by combing a detection model with a segmentation model. Both models are based on the U-Net architecture. With the proposed method, we achieve very promising segmentation results on our T1-weighted MR image dataset collected from rectal cancer patients. T1-weighted images are useful for the radiologist to observe the anatomical information. Therefore, this work has the potential to be further developed to help clinicians diagnose rectal cancer and design treatment plans. This study is a proof-of-concept that introducing detection to segmentation is effective. In the future, we will investigate more on the individual models.Acknowledgements
No acknowledgement found.References
1. Kim Y H, Kim D Y, Kim T H, et al. Usefulness of magnetic resonance volumetric evaluation in predicting response to preoperative concurrent chemoradiotherapy in patients with resectable rectal cancer. International Journal of Radiation Oncology Biology Physics, 2005;62(3), 761-768.
2. Pizzi A D, Basilico R, Cianci R, et al. Rectal cancer MRI: protocols, signs and future perspectives radiologists should consider in everyday clinical practice. Insights into imaging. 2018;9(4), 405-412.
3. Ronneberger O, Fischer P, et al. U-net: Convolutional networks for biomedical image segmentation. Proc. MICCAI. 2015;234-241.
4. Badrinarayanan V, Kendall A, Cipolla R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE transactions on pattern analysis and machine intelligence, 2017;39(12): 2481-2495.
5. Liu W, Anguelov D, Erhan D, et al. Ssd: Single shot multibox detector. Proc. European conference on computer vision. 2016;21-37
6. Trebeschi S, van Griethuysen J J, Lambregts D M, et al. Deep learning for fully-automated localization and segmentation of rectal cancer on multiparametric MR. Scientific reports. 2017;7(1), 5301.
7. Kim J, Oh J E, Lee J, et al. Rectal cancer: Toward fully automatic discrimination of T2 and T3 rectal cancers using deep convolutional neural network. International Journal of Imaging Systems and Technology. 2019;29(3), 247-259.
8. Wang M, Xie P, Ran Z, et al. Full convolutional network based multiple side‐output fusion architecture for the segmentation of rectal tumors in magnetic resonance images: A multi‐vendor study. Medical physics. 2019;46(6): 2659-2668
Figures