3517
Segmenting Brain Tumor Lesion from 3D FLAIR MR Images using Support Vector Machine approach1Center for Biomedical Engineering, Indian Institute of Technology, Delhi, India, 2Fortis Memorial Research Institute, Gurugram, India, 3Electrical Engineering, Indian Institute of Technology, Delhi, India, 4Biomedical Engineering, AIIMS, New Delhi, India
Synopsis
Segmentation of brain tumor lesion is important for diagnosis and treatment planning. Tumor tissue and edema usually appears hyperintense on fluid-attenuated-inversion-recovery (FLAIR) MR images. FLAIR images are widely used for brain tumor localization and segmentation purpose. In this study, a Support-Vector-Machine (SVM) model was developed for segmentation of FLAIR hyper-intense region semi-automatically using BraTS 2018 dataset. The proposed approach require a minimal user involvement in selecting one region around tumor in the central slice. It was observed that proposed SVM approach segmentation results shows better dice coefficient in comparison to what reported in literature.
Introduction
Accurate detection and segmentation of brain tumors in MRI images assists in treatment planning. Increasing amount of brain imaging data has made the clinical expert job tiresome1. To reduce the manual effort in accurate diagnosis by the clinician and speedup the process, several alternative ways has been investigated1-8. Various semi-automated and fully automated process have been proposed in literature to carryout tumor segmentation task using MRI images1-8. In clinical practice, semi-automatic approaches are adopted2. Advantage of semi-automatic approach is, radiologist control over segmentation process to ensure accuracy2. This study aims to develop a machine learning (i.e. support vector machine) based semi-automatic system to carry out the tumor segmentation task from 3D Fluid-attenuated inversion recovery (FLAIR) MR images of brain.Method
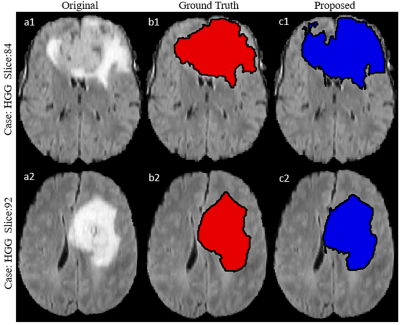
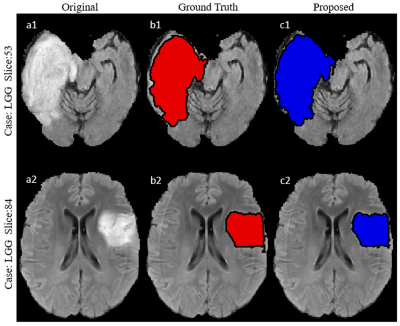
A total 60 FLAIR images (30 images from LGG and 30 images from HGG) were chosen from BraTS dataset for development and testing of SVM model9-11. Intensity value of each voxel of FLAIR images is only considered feature. For training SVM Model, a rough Region of Interest (ROI) of any shape (square, polygon, circle etc.) is selected on FLAIR image which contains tumor. All futher computation was performed on this selected ROI instead on whole FLAIR image. Intensity scaling (0-255) was performed. Let’s represent this ROI mask as R1mask (Figure 1). Segmented tumor masks are also available in BraTS 2018 dataset, which serve as ground-truth for training and validation. Let’s represent this ground-truth mask, over same ROI, as R2mask(Figure 1). Multiply R1mask with R2mask provided hyperintense region of FLAIR image and let’s represent it by R3. Subtract R3 from R1mask to obtain Rsub. From Rsub a data set V1- is created for non-zeros intensity values containing two column vectors: intensity value and label (figure 1). Put all labels ‘-1’ in label field of data set V1- . Similarly generate a data set V1+ of similar structure from R3. Put all labels ‘1’ in label field of vector V1+ . This process is carried out for 20 FLAIR images (10 images from each HGG and LGG respectively) containing tumor region. Finally, a combined dataset V is created containing all intensities along with their corresponding labels i.e. 1 or -1. Every voxel intensity along with their corresponding labels was considered as sample. Dataset was divided into training set and testing set in 90:10 ratio. Ten-fold cross validation was used to validate proposed SVM model. SVM model was trained using linear kernel. A range of C (0.0001-1000) in SVM model was investigated to obtain the value of C for which mean of Ten-fold cross validation error was minimum. Observed C value for which model produce minimum Ten-fold cross validation error was accepted to develop the final model for FLAIR hyperintense lesion segmentation. To use this final developed model, a user has to visually scan the FLAIR images of a particular patient in search of images containing tumor. A FLAIR image which contain tumor of larger size compared to other tumor slices is chosen and around tumor, a rough ROI is drawn on this chosen FLAIR image and applied to all surrounding slices automatically. From these ROIs, proposed SVM model will result tumor segmentation for all FLAIR images which contains tumor. For testing accuracy of segmentation, dice coefficient index was computed between proposed segmentation based mask and available groundtruth mask from data of new test patients.Result
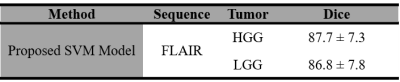
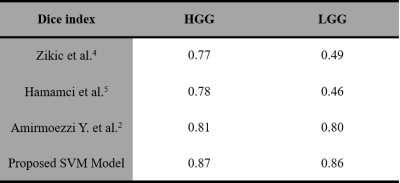
Observed error percentage in training, validation and testing set were 8.5973, 8.594 and 8.597 respectively for best C (equal to 1). Developed SVM model, tested against 40 images (HGG=20, LGG=20) taken from BraTS 2018 dataset provided an average dice score against ground truth (available in BraTS 2018 dataset), equal to 87.7 and 86. 8 for HGG and LGG images respectively (Table I). Proposed model resulted better segmentation accuracy compared to previously reported results available in literature (Table II).Discussion
Advantage of using BraTS 2018 dataset is that it contains image sequences which were collected worldwide by using different clinical image acquisition protocol, different scanners and from multiple organizations. It is believe that Training SVM model on such a dataset generalizes SVM model. A manual ROI is selected around specific region of interest (around tumor) of any shape. Selecting rough ROI (around tumor) did not requires much expertise and skills. As all the processing is performed on selected ROI on FLAIR image instead on whole image, resulted in reduced computation time. Voxel intensity is only considered feature in training SVM model at this stage for classifying pixel as tumor or non-tumor pixel. Proposed approach not only exploits the advantage of semi-automatic approaches but also take care interobserver variability in segmentation process.Conclusion
In this study, an effort has been made to develop a semi-automatic model (SVM based) to segment hyperintense lesion in FLAIR images for clinical use. The proposed model uses single imaging sequence (FLAIR), an ROI which is smaller in size as compared to the original image and one feature i.e. voxel intensity, which makes it relatively fast. It was found that developed SVM model when tested against some other images which were not included in the training (from BraTS 2018 dataset) showed improved performance compared to other approach’s results reported in literature.Acknowledgements
This work was supported by Indian Institute of Technology Delhi and Fortis Memorial Research Institute Gurugram. The authors thanks Dr. Anirban Sengupta, Dr. Ayan, Rafeek, Esha, Dharmesh, Umang and Dr. Sumantra Dutta Roy for their valuable suggestion.References
1. Mohan G, Monica Subashini M. MRI based medical image analysis: Survey on brain tumor grade classification. Biomedical Signal Processing and Control (2018); volume 39, pages 139-161.
2. Amirmoezzi, Y., Salehi, S., Parsaei, H. et al. A knowledge-based system for brain tumor segmentation using only 3D FLAIR images. Australas Phys Eng Sci Med (2019) 42: 529.
3. Korfiatis P, Kline TL, Erickson BJ. Automated Segmentation of Hyperintense Regions in FLAIR MRI Using Deep Learning. Tomography. 2016; 2(4):334–340.
4. Zikic D, Glocker B, Konukoglu E, Shotton J, Criminisi A, Ye DH, et al. Context-sensitive classification forests for segmentation of brain tumor tissues. In: MICCAI 2012 Chall Multimodal Brain Tumor Segmentation
5. Hamamci A, Kucuk N, Karaman K, Engin K, Unal G. Tumor-cut: segmentation of brain tumors on contrast enhanced MR images for radiosurgery applications. IEEE Trans Med Imaging.2012; 31:790–804
6. Schmidt P, Pongratz V et al. Automated segmentation of changes in FLAIR-hyperintense white matter lesions in multiple sclerosis on serial magnetic resonance imaging. NeuroImage: Clinical2019; 2213-1582.
7. Sengupta A, Agarwal S, Gupta PK, et al. On differentiation between vasogenic edema and non-enhancing tumor in high-grade glioma patients using a support vector machine classifier based upon pre and post-surgery MRI images. Eur J Radiol. (2018),106:199-208.
8. Duong M.T., Rudie J.D., et al. Convolutional Neural Network for Automated FLAIR Lesion Segmentation on Clinical Brain MR Imaging. American Journal of Neuroradiology. 2019.
9. Menze B. H., Jakab A., et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Transactions on Medical Imaging. 2015; 34(10), 1993-2024.
10. Bakas S., Akbari H., et al. Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Nature Scientific Data. 201; 4:170117.
11. Bakas S., Akbari H., et al. Identifying the Best Machine Learning Algorithms for Brain Tumor Segmentation, Progression Assessment, and Overall Survival Prediction in the BRATS Challenge. 2018; arXiv preprint arXiv:1811.02629.
Figures